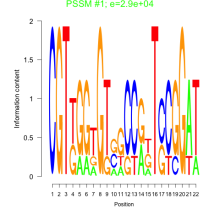
Phatr_bicluster_0188 Residual: 0.15
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0188 | 0.15 | Phaeodactylum tricornutum |
Displaying 1 - 20 of 20
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12732 adenylate kinase cytosolic (adk)
PHATRDRAFT_13261 at4g39970 t5j17_140 (PLN02779)
PHATRDRAFT_18337 glutamine-dependent asparagine synthetase (PLN02549)
PHATRDRAFT_21388 (COG1204)
PHATRDRAFT_26813 homoserine dehydrogenase (ThrA)
PHATRDRAFT_32202 lysyl-trna synthetase (lysS)
PHATRDRAFT_34747 n-anthranilate isomerase (PRAI)
PHATRDRAFT_34933 peptidylprolyl isomeraselike (cyclophilin_ABH_like)
PHATRDRAFT_41721 cpn60 protein (groEL)
PHATRDRAFT_45306 (PLDc_SF superfamily)
PHATRDRAFT_45434 at5g28840 f7p1_20 (PLN02695)
PHATRDRAFT_46281 cg9915-isoform b
PHATRDRAFT_46333 heat shock protein 70 (HSP70_NBD)
PHATRDRAFT_46438 cytochrome p450-like protein (p450 superfamily)
PHATRDRAFT_46599 clathrin light chain (Clathrin_lg_ch superfamily)
PHATRDRAFT_54590 (Tic22 superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006430 | lysyl-tRNA aminoacylation | 0.00641182 | 1 | Phatr_bicluster_0188 |
| GO:0006529 | asparagine biosynthesis | 0.0127857 | 1 | Phatr_bicluster_0188 |
| GO:0006422 | aspartyl-tRNA aminoacylation | 0.0191218 | 1 | Phatr_bicluster_0188 |
| GO:0006568 | tryptophan metabolism | 0.0191218 | 1 | Phatr_bicluster_0188 |
| GO:0009225 | nucleotide-sugar metabolism | 0.0222757 | 1 | Phatr_bicluster_0188 |
| GO:0008652 | amino acid biosynthesis | 0.0222757 | 1 | Phatr_bicluster_0188 |
| GO:0044267 | cellular protein metabolism | 0.0316815 | 1 | Phatr_bicluster_0188 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 0.0347981 | 1 | Phatr_bicluster_0188 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.154889 | 1 | Phatr_bicluster_0188 |
| GO:0008152 | metabolism | 0.242573 | 1 | Phatr_bicluster_0188 |


Comments