Thaps_bicluster_0099 Residual: 0.44
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0099 | 0.44 | Thalassiosira pseudonana |
Displaying 1 - 30 of 30
" class="views-fluidgrid-wrapper clear-block">
1053 hypothetical protein
18795 (Tp_Myb3R7) SANT
19345 HepA
1942 hypothetical protein
1975 hypothetical protein
22484 hypothetical protein
22642 SET
23000 hypothetical protein
23054 hypothetical protein
23561 hypothetical protein
23773 hypothetical protein
25324 hypothetical protein
25621 hypothetical protein
2774 hypothetical protein
29304 (TUB1) PLN00221
3279 hypothetical protein
3537 hypothetical protein
3649 hypothetical protein
36522 (TOP2) PLN03128
36788 PLN02438
3765 hypothetical protein
38187 Peptidase_C14
38732 BaeS
38948 Peptidase_C97
6104 hypothetical protein
6408 hypothetical protein
6957 hypothetical protein
7036 WD40 superfamily
9238 hypothetical protein
9414 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006021 | myo-inositol biosynthesis | 0.00454357 | 1 | Thaps_bicluster_0099 |
| GO:0051258 | protein polymerization | 0.0180619 | 1 | Thaps_bicluster_0099 |
| GO:0006265 | DNA topological change | 0.0225308 | 1 | Thaps_bicluster_0099 |
| GO:0006259 | DNA metabolism | 0.0314132 | 1 | Thaps_bicluster_0099 |
| GO:0008654 | phospholipid biosynthesis | 0.0314132 | 1 | Thaps_bicluster_0099 |
| GO:0009401 | phosphoenolpyruvate-dependent sugar phosphotransferase system | 0.0402221 | 1 | Thaps_bicluster_0099 |
| GO:0007160 | cell-matrix adhesion | 0.0402221 | 1 | Thaps_bicluster_0099 |
| GO:0007155 | cell adhesion | 0.0662137 | 1 | Thaps_bicluster_0099 |
| GO:0007018 | microtubule-based movement | 0.0936478 | 1 | Thaps_bicluster_0099 |
| GO:0006412 | protein biosynthesis | 0.319237 | 1 | Thaps_bicluster_0099 |

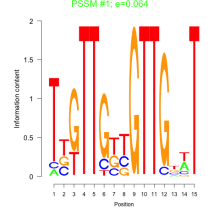

Comments