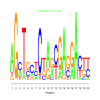
Module 226 Residual: 0.52
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0226 | v02 | 0.52 | -11.21 |
| Rv0194 ATP binding cassette (ABC) transporter homolog | Rv0561c Possible oxidoreductase (EC 1.-.-.-) |
| Rv0772 Phosphoribosylamine--glycine ligase (EC 6.3.4.13) | Rv0819 Acetyl-CoA:Cys-GlcN-Ins acetyltransferase, mycothiol synthase MshD |
| Rv0918 | Rv0919 GCN5-related N-acetyltransferase; Histone acetyltransferase HPA2 and related acetyltransferases |
| Rv1272c Lipid A export ATP-binding/permease protein MsbA | Rv1273c ABC transporter ATP-binding protein |
| Rv1666c Cytochrome P450 139 | Rv1718 |
| Rv1979c Amino acid permease | Rv2435c Adenylate cyclase (EC 4.6.1.1) |
| Rv0944 Formamidopyrimidine-DNA glycosylase (EC 3.2.2.23) | Rv0945 Oxidoreductase, short-chain dehydrogenase/reductase family (EC 1.1.1.-) |
| Rv2434c Small-conductance mechanosensitive channel membrane protein | Rv3542c Conserved protein IgrD |
|
GO:0005618 | cell wall |
|
GO:0005886 | plasma membrane |
|
GO:0015562 | efflux transmembrane transporter activity |
|
GO:0004637 | phosphoribosylamine-glycine ligase activity |
|
GO:0040007 | growth |
|
GO:0008080 | N-acetyltransferase activity |
|
GO:0006950 | response to stress |
|
GO:0010125 | mycothiol biosynthetic process |
|
GO:0035447 | mycothiol synthase activity |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0052572 | response to host immune response |
|
GO:0004016 | adenylate cyclase activity |
|
GO:0004519 | endonuclease activity |
|
GO:0006281 | DNA repair |
|
GO:0006289 | nucleotide-excision repair |
|
GO:0034599 | cellular response to oxidative stress |
|
GO:0044117 | growth of symbiont in host |


















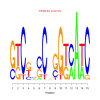
Discussion