Module 316 Residual: 0.52
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0316 | v02 | 0.52 | -9.15 |
| Rv1435c \Probable conserved Proline, Glycine, Valine-rich secreted protein\\\"" | Rv1566c Invasion protein |
| Rv2145c FIG055075: Possibly a cell division protein, antigen 84 in Mycobacteria | Rv2165c rRNA small subunit methyltransferase H |
| Rv2166c Cell division protein MraZ | Rv2525c Putative secreted protein |
| Rv2986c DNA-binding protein HU / low-complexity, AKP-rich domain | Rv3246c DNA-binding response regulator mtrA |
| Rv3258c | Rv3413c |
| Rv3414c RNA polymerase sigma-70 factor | Rv3587c |
| Rv0430 | Rv0660c Prevent host death protein, Phd antitoxin |
| Rv0883c | Rv1497 Esterase LipL |
| Rv2927c Cell division initiation protein | Rv3244c LpqB |
| Rv3245c Sensor histidine kinase MtrB (EC 2.7.3.-) | Rv3260c Sporulation regulatory protein WhiB |
| Rv3415c |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0009165 | GO:1901293 | GO:0009117 | GO:0016742 | nucleotide biosynthetic process | nucleoside phosphate biosynthetic proces... | nucleotide metabolic process | hydroxymethyl-, formyl- and related tran... |
|
GO:0005576 | extracellular region |
|
GO:0005515 | protein binding |
|
GO:0005618 | cell wall |
|
GO:0005886 | plasma membrane |
|
GO:0008360 | regulation of cell shape |
|
GO:0009273 | peptidoglycan-based cell wall biogenesis |
|
GO:0040007 | growth |
|
GO:0050821 | protein stabilization |
|
GO:0060187 | cell pole |
|
GO:0046677 | response to antibiotic |
|
GO:0008610 | lipid biosynthetic process |
|
GO:0003677 | DNA binding |
|
GO:0010106 | cellular response to iron ion starvation |
|
GO:0008150 | biological_process |
|
GO:0006468 | protein phosphorylation |
|
GO:0044117 | growth of symbiont in host |
|
GO:0045893 | positive regulation of transcription, DNA-dependent |
|
GO:0009405 | pathogenesis |
|
GO:0009408 | response to heat |
|
GO:0016987 | sigma factor activity |
|
GO:0046688 | response to copper ion |
|
GO:0004806 | triglyceride lipase activity |
|
GO:0005829 | cytosol |























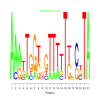
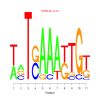
Discussion