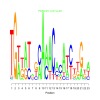
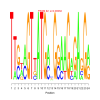
Module 331 Residual: 0.41
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0331 | v02 | 0.41 | -17.24 |
| Rv0046c Inositol-1-phosphate synthase (EC 5.5.1.4) | Rv0145 |
| Rv0146 | Rv0244c 3-methylmercaptopropionyl-CoA dehydrogenase (DmdC) |
| Rv0483 Possible lipoprotein LprQ | Rv1778c |
| Rv1779c | Rv2428 Alkyl hydroperoxide reductase protein C (EC 1.6.4.-) |
| Rv2429 Alkylhydroperoxidase protein D | Rv2518c Lipoprotein LppS |
| Rv2699c | Rv2707 Ribonuclease BN (EC 3.1.-.-) |
| Rv2724c Acyl-CoA dehydrogenase, short-chain specific (EC 1.3.99.2) | Rv3484 Cell envelope-associated transcriptional attenuator LytR-CpsA-Psr, subfamily A1 (as in PMID19099556) |
| Rv3567c Nitrilotriacetate monooxygenase component B (EC 1.14.13.-) | Rv3571 Phenylacetate-CoA oxygenase/reductase, PaaK subunit |
| Rv3570c POSSIBLE OXIDOREDUCTASE |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0044238 | GO:0044237 | GO:0009987 | GO:0071704 | GO:0005623 | GO:0044464 | GO:0015934 | GO:0015935 | primary metabolic process | cellular metabolic process | cellular process | organic substance metabolic process | cell | cell part | large ribosomal subunit | small ribosomal subunit |
|
GO:0004512 | inositol-3-phosphate synthase activity |
|
GO:0008270 | zinc ion binding |
|
GO:0009405 | pathogenesis |
|
GO:0005829 | cytosol |
|
GO:0005886 | plasma membrane |
|
GO:0005576 | extracellular region |
|
GO:0052572 | response to host immune response |
|
GO:0004601 | peroxidase activity |
|
GO:0005515 | protein binding |
|
GO:0008785 | alkyl hydroperoxide reductase activity |
|
GO:0032843 | hydroperoxide reductase activity |
|
GO:0045454 | cell redox homeostasis |
|
GO:0051260 | protein homooligomerization |
|
GO:0051409 | response to nitrosative stress |
|
GO:0051920 | peroxiredoxin activity |
|
GO:0055114 | oxidation-reduction process |
|
GO:0042803 | protein homodimerization activity |
|
GO:0070207 | protein homotrimerization |
|
GO:0005618 | cell wall |
|
GO:0046812 | host cell surface binding |
|
GO:0001666 | response to hypoxia |
|
GO:0044117 | growth of symbiont in host |



















Discussion