Module 177 Residual: 0.50
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0177 | v02 | 0.50 | -14.95 |
| Rv1683 Polyhydroxyalkanoic acid synthase | Rv1708 Chromosome (plasmid) partitioning protein ParA / Sporulation initiation inhibitor protein Soj |
| Rv1709 Segregation and condensation protein A | Rv1710 Segregation and condensation protein B |
| Rv1711 Ribosomal small subunit pseudouridine synthase A (EC 4.2.1.70) | Rv1712 Cytidylate kinase (EC 2.7.4.14) |
| Rv1713 GTP-binding protein EngA | Rv2426c MoxR-like ATPases |
| Rv2604c Pyridoxine biosynthesis glutamine amidotransferase, glutaminase subunit (EC 2.4.2.-) | Rv3226c |
| Rv3227 5-Enolpyruvylshikimate-3-phosphate synthase (EC 2.5.1.19) | Rv3228 Ribosome small subunit-stimulated GTPase EngC |
| Rv1252c Putative lipoprotein lprE precursor | Rv2857c Short-chain dehydrogenase/reductase in hypothetical Actinobacterial gene cluster |
| Rv2858c Aldehyde dehydrogenase in hypothetical Actinobacterial gene cluster | Rv2859c COG2071: predicted glutamine amidotransferases in hypothetical Actinobacterial gene cluster |
| Rv2860c | Rv3106 Ferredoxin--NADP(+) reductase, actinobacterial (eukaryote-like) type (EC 1.18.1.2) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006541 | glutamine metabolic process |
|
GO:0008415 | NA |
|
GO:0042619 | poly-hydroxybutyrate biosynthetic process |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0004127 | cytidylate kinase activity |
|
GO:0005525 | GTP binding |
|
GO:0005622 | intracellular |
|
GO:0005618 | cell wall |
|
GO:0003866 | 3-phosphoshikimate 1-carboxyvinyltransferase activity |
|
GO:0005576 | extracellular region |
|
GO:0004029 | aldehyde dehydrogenase (NAD) activity |
|
GO:0004049 | anthranilate synthase activity |
|
GO:0004356 | glutamate-ammonia ligase activity |
|
GO:0051260 | protein homooligomerization |
|
GO:0004324 | ferredoxin-NADP+ reductase activity |
|
GO:0008860 | ferredoxin-NAD+ reductase activity |
|
GO:0050660 | flavin adenine dinucleotide binding |
|
GO:0055114 | oxidation-reduction process |
|
GO:0070401 | NADP+ binding |




















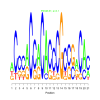
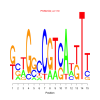
Discussion