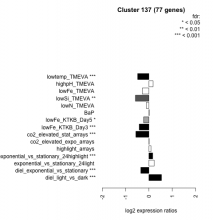
Thaps_hclust_0137 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006118 | electron transport | 0.0194004 | 1 | Thaps_hclust_0137 |
| GO:0015780 | nucleotide-sugar transport | 0.0296321 | 1 | Thaps_hclust_0137 |
| GO:0019538 | protein metabolism | 0.0305249 | 1 | Thaps_hclust_0137 |
| GO:0006094 | gluconeogenesis | 0.0527255 | 1 | Thaps_hclust_0137 |
| GO:0006342 | chromatin silencing | 0.0527255 | 1 | Thaps_hclust_0137 |
| GO:0006099 | tricarboxylic acid cycle | 0.0584155 | 1 | Thaps_hclust_0137 |
| GO:0006006 | glucose metabolism | 0.0584155 | 1 | Thaps_hclust_0137 |
| GO:0008643 | carbohydrate transport | 0.080847 | 1 | Thaps_hclust_0137 |
| GO:0016192 | vesicle-mediated transport | 0.0918683 | 1 | Thaps_hclust_0137 |
| GO:0006725 | aromatic compound metabolism | 0.124172 | 1 | Thaps_hclust_0137 |
|
1038 : SPRYD7 |
5212 : (Tp_CCCH26) regulator [Rayko] |
11184 : hypothetical protein |
25535 : hypothetical protein |
|
1762 : PRA1 superfamily |
5225 : Nsp1_C superfamily |
11187 : hypothetical protein |
25719 : hypothetical protein |
|
1850 : PKc_like superfamily |
5335 : Cyt_b561 superfamily |
11576 : MPP_Vps29 |
25860 : hypothetical protein |
|
1871 : hypothetical protein |
5420 : hypothetical protein |
11756 : SURF1 superfamily |
26769 : ERG4_ERG24 superfamily |
|
1912 : 2OG-FeII_Oxy_3 superfamily |
5600 : Thioredoxin_like superfamily |
17314 : Solute_trans_a superfamily |
30149 : Solute_trans_a |
|
2086 : zf-UBP |
5659 : hypothetical protein |
20680 : hypothetical protein |
32945 : (SDH1) sdhB |
|
2107 : hypothetical protein |
6032 : Ubox |
20977 : COG0398 |
33024 : cysteine_hydrolases superfamily |
|
2319 : hypothetical protein |
6140 : hypothetical protein |
21083 : hypothetical protein |
34514 : PLN02539 |
|
2515 : nst superfamily |
6594 : hypothetical protein |
21168 : hypothetical protein |
35956 : SIR2 superfamily |
|
3064 : hypothetical protein |
6710 : hypothetical protein |
21681 : hypothetical protein |
37584 : Cupin_3 |
|
3167 : hypothetical protein |
7696 : 2_5_RNA_ligase2 |
21690 : hypothetical protein |
37793 : hypothetical protein |
|
3204 : hypothetical protein |
8821 : CHD |
21713 : UbiH |
38602 : Ndh |
|
3426 : zf-DNL superfamily |
8841 : P4Hc |
21887 : hypothetical protein |
38764 : Bromodomain |
|
3661 : MMCE |
9793 : NADB_Rossmann superfamily |
22307 : TRAPP |
261394 : Aldo_ket_red |
|
3913 : LCM |
10059 : hypothetical protein |
22682 : hypothetical protein |
263399 : CypX |
|
3966 : PT_UbiA superfamily |
10353 : hypothetical protein |
23082 : hypothetical protein |
263810 : PTZ00430 |
|
4114 : GAT_SF |
10624 : hypothetical protein |
24245 : hypothetical protein |
264157 : CBS_pair superfamily |
|
4456 : Abhydrolase_6 |
10856 : hypothetical protein |
24480 : ECM4 |
269113 : M3_like |
|
5038 : DUF179 |
10889 : hypothetical protein |
25506 : hypothetical protein |
270302 : hypothetical protein |
|
5094 : hypothetical protein |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | 0.540 | 0.000485 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | -0.413 | 0.000862 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | -0.226 | 0.0236 |
| BaP | BaP | 0.015 | 0.915 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | 0.162 | 0.000526 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | -0.560 | 0.000658 |
| lowtemp_TMEVA | lowtemp_TMEVA | -0.484 | 0.000735 |
| highpH_TMEVA | highpH_TMEVA | 0.179 | 0.0506 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.011 | 0.985 |
| lowFe_TMEVA | lowFe_TMEVA | -0.285 | 0.053 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.225 | 0.263 |
| lowN_TMEVA | lowN_TMEVA | -0.113 | 0.761 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | -0.349 | 0.000602 |
| lowSi_TMEVA | lowSi_TMEVA | -0.578 | 0.00135 |
| highlight_arrays | highlight_arrays | 0.091 | 0.326 |