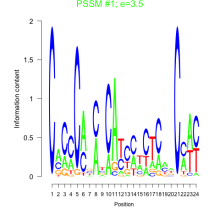
Thaps_bicluster_0032 Residual: 0.47
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0032 | 0.47 | Thalassiosira pseudonana |
Displaying 1 - 57 of 57
" class="views-fluidgrid-wrapper clear-block">
10024 hypothetical protein
10027 hypothetical protein
10128 hypothetical protein
10194 hypothetical protein
10598 hypothetical protein
10772 hypothetical protein
11068 (PSA6) proteasome_alpha_type_6
11278 hypothetical protein
11357 hypothetical protein
11409 hypothetical protein
11618 hypothetical protein
11683 hypothetical protein
11993 CGS_like
12131 hypothetical protein
12206 Bestrophin superfamily
14555 COG0523
19598 Tryp_SPc
2037 hypothetical protein
22976 PLN03199
23566 hypothetical protein
23596 TPR
24057 hypothetical protein
24418 hypothetical protein
25241 hypothetical protein
25467 hypothetical protein
2552 DUS_like_FMN
2600 hypothetical protein
261453 (Tp_Hox8) regulator [Rayko]
261803 LRR_RI superfamily
262180 hypothetical protein
263510 TOPRIM_TopoIIB_SPO
3056 hypothetical protein
31200 DSPc superfamily
33676 DUF846
33984 PKc_like superfamily
35381 hypothetical protein
37683 ANK
38153 B9-C2
4293 hypothetical protein
5180 hypothetical protein
5272 hypothetical protein
5580 hypothetical protein
5855 AAA_12
6294 hypothetical protein
6360 hypothetical protein
6576 hypothetical protein
7199 hypothetical protein
7292 hypothetical protein
7375 hypothetical protein
7416 hypothetical protein
7953 AIM24
8111 VIT1
8994 hypothetical protein
913 hypothetical protein
9310 hypothetical protein
9330 hypothetical protein
9602 ATPase-IIIA_H
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006265 | DNA topological change | 0.0265779 | 1 | Thaps_bicluster_0032 |
| GO:0006636 | fatty acid desaturation | 0.0318146 | 1 | Thaps_bicluster_0032 |
| GO:0006259 | DNA metabolism | 0.0370252 | 1 | Thaps_bicluster_0032 |
| GO:0008033 | tRNA processing | 0.047369 | 1 | Thaps_bicluster_0032 |
| GO:0006520 | amino acid metabolism | 0.0550594 | 1 | Thaps_bicluster_0032 |
| GO:0051341 | regulation of oxidoreductase activity | 0.0601545 | 1 | Thaps_bicluster_0032 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0677496 | 1 | Thaps_bicluster_0032 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.128874 | 1 | Thaps_bicluster_0032 |
| GO:0006508 | proteolysis and peptidolysis | 0.179105 | 1 | Thaps_bicluster_0032 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.197871 | 1 | Thaps_bicluster_0032 |


Comments