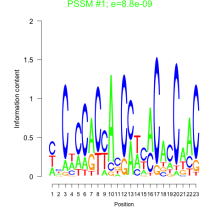
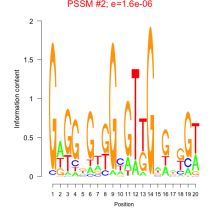
Thaps_bicluster_0162 Residual: 0.37
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0162 | 0.37 | Thalassiosira pseudonana |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
22139 Cyt_C5_DNA_methylase superfamily
22808 SpoU_methylase superfamily
23175 hypothetical protein
23179 PTZ00416
24552 hypothetical protein
24786 hypothetical protein
263424 PLN02551
263630 (MTS) tRNA_bind_EMAP-II_like
26403 PLN03233
264164 Porin3_Tom40
267957 translation elongation factor ef-1 alpha
268714 TypA_BipA
268807 (RIR1_2) PLN02437
269018 hypothetical protein
269371 hypothetical protein
269399 PLN02678
269616 PLN03127
27997 (DAD1) PRK00911
28330 PurH
3007 hypothetical protein
31809 adk
31929 (GGPS5) Trans_IPPS_HT
36339 (ITS1) PLN02882
41952 EF-G
42213 PLN02286
42226 PLN03233
9318 tsf
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006418 | tRNA aminoacylation for protein translation | 0.000182366 | 0.499135 | Thaps_bicluster_0162 |
| GO:0006414 | translational elongation | 0.00164003 | 1 | Thaps_bicluster_0162 |
| GO:0006412 | protein biosynthesis | 0.00962997 | 1 | Thaps_bicluster_0162 |
| GO:0009082 | branched chain family amino acid biosynthesis | 0.0103113 | 1 | Thaps_bicluster_0162 |
| GO:0006420 | arginyl-tRNA aminoacylation | 0.0103113 | 1 | Thaps_bicluster_0162 |
| GO:0006428 | isoleucyl-tRNA aminoacylation | 0.0103113 | 1 | Thaps_bicluster_0162 |
| GO:0006434 | seryl-tRNA aminoacylation | 0.0154287 | 1 | Thaps_bicluster_0162 |
| GO:0006424 | glutamyl-tRNA aminoacylation | 0.0205206 | 1 | Thaps_bicluster_0162 |
| GO:0006164 | purine nucleotide biosynthesis | 0.0205206 | 1 | Thaps_bicluster_0162 |
| GO:0006820 | anion transport | 0.0406365 | 1 | Thaps_bicluster_0162 |

Comments