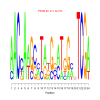
Module 294 Residual: 0.47
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0294 | v02 | 0.47 | -13.41 |
| Rv1368 Lipoprotein LprF | Rv1829 |
| Rv1891 Possible membrane protein | Rv1892 Possible membrane protein |
| Rv1893 | Rv2348c |
| Rv2406c Inosine-5'-monophosphate dehydrogenase (EC 1.1.1.205) | Rv2485c Carboxylesterase LipQ |
| Rv2632c | Rv2633c |
| Rv3612c | Rv3613c |
| Rv3614c ESX-1 secretion system protein (Rv3614c) | Rv3615c Highly immunodominant RD1 (Region of Difference 1)-dependent secreted antigen specific for Mycobacterium tuberculosis infection |
| Rv3616c ESX-1 secretion system protein (Rv3616c) | Rv3633 Protein involved in biosynthesis of mitomycin antibiotics/polyketide fumonisin |
| Rv1904 Anti-sigma F factor antagonist (spoIIAA-2); Anti-sigma B factor antagonist RsbV | Rv2459 Multidrug resistance integral membrane efflux protein EmrB |
| Rv3750c Excisionase |
|
GO:0005515 | protein binding |
|
GO:0005886 | plasma membrane |
|
GO:0005576 | extracellular region |
|
GO:0009405 | pathogenesis |
|
GO:0003938 | IMP dehydrogenase activity |
|
GO:0005829 | cytosol |
|
GO:0042783 | active evasion of host immune response |





















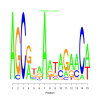
Discussion