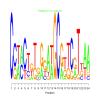
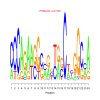
Module 467 Residual: 0.48
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0467 | v02 | 0.48 | -17.47 |
| Rv0121c PPOX class F420-dependent enzyme | Rv0327c Cytochrome P450 135A1 |
| Rv0328 HTH-type transcriptional regulator BetI | Rv0329c |
| Rv0378 PE_PGRS family protein | Rv0522 Aromatic amino acid transport protein AroP |
| Rv0616c | Rv1358 Adenylyl cyclase class-3/4/guanylyl cyclase / Disease resistance domain-containing protein / Tetratricopeptide repeat-containing protein / Transcriptional regulator, LuxR family |
| Rv1359 Adenylyl cyclase class-3/4/guanylyl cyclase | Rv0235c Possible membrane protein |
| Rv0376c CO dehydrogenases maturation factor, CoxF family | Rv0377 Transcription regulator in CO-DH cluster |
| Rv1123c Possible peroxidase bpoB (Non-haem peroxidase) (EC 1.11.1.-) | Rv1124 Probable epoxide hydrolase ephC (EC 3.3.2.9) |
| Rv1125 | Rv2943 Mobile element protein |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006091 | GO:0055114 | GO:0044710 | GO:0019239 | GO:0016814 | generation of precursor metabolites and ... | oxidation-reduction process | single-organism metabolic process | deaminase activity | hydrolase activity, acting on carbon-nit... |
|
GO:0005886 | plasma membrane |
|
GO:0043091 | L-arginine import |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |


















Discussion