Module 523 Residual: 0.52
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0523 | v02 | 0.52 | -5.29 |
| Rv2982c Glycerol-3-phosphate dehydrogenase [NAD(P)+] (EC 1.1.1.94) | Rv0057 |
| Rv0058 Replicative DNA helicase (EC 3.6.1.-) @ intein-containing | Rv0059 |
| Rv0060 ADP-ribose 1-phosphate phophatase related protein | Rv0134 epoxide hydrolase |
| Rv0743c | Rv0744c POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| Rv0962c lipoprotein, putative | Rv1017c Ribose-phosphate pyrophosphokinase (EC 2.7.6.1) |
| Rv1920 Acyltransferase family protein | Rv2716 Phenazine biosynthesis protein PhzF like |
| Rv2734 Phage Gp37Gp68 protein | Rv3336c Tryptophanyl-tRNA synthetase (EC 6.1.1.2) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0031177 | GO:0072341 | GO:0048037 | GO:0033218 | GO:0016597 | phosphopantetheine binding | modified amino acid binding | cofactor binding | amide binding | amino acid binding |
|
GO:0047952 | glycerol-3-phosphate dehydrogenase [NAD(P)+] activity |
|
GO:0005886 | plasma membrane |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |
|
GO:0006974 | response to DNA damage stimulus |
|
GO:0040007 | growth |
|
GO:0004749 | ribose phosphate diphosphokinase activity |
|
GO:0004830 | tryptophan-tRNA ligase activity |
















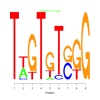

Discussion