Citation: Arrieta-Ortiz et al. 2020. bioRxiv. Predictive regulatory and metabolic network models for systems analysis of Clostridioides difficile
Clostridioides difficile causes >500,000 infections, >30,000 deaths, and > $5 billion/year in US healthcare costs, and rates continue to rise. Antimicrobial therapy that ablates the commensal microbiota commonly triggers infection, allowing the pathogen to proliferate and release toxins that damage host mucosal surfaces. Resistance to commonly prescribed antibiotics occurs in >20% of patient isolates, a problem that confounds treatment and increases risks for recurrent infections. In this research program, we are building predictive and mechanistic models to define mechanisms by which C. difficile responds to antibiotics, host and microbiota-origin factors, and to develop therapeutic interventions to prevent colonization, infection and recurrence of C. difficile.
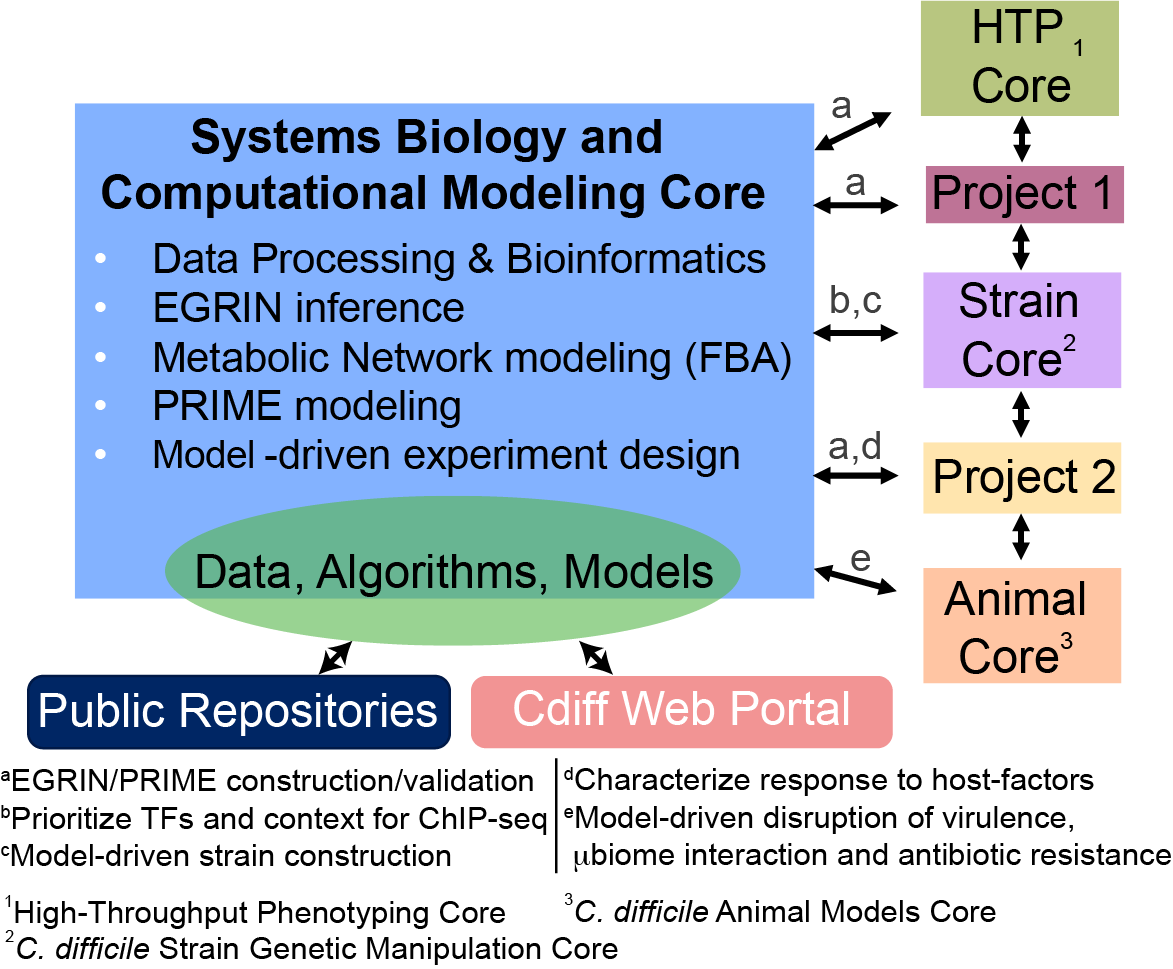
The multi-institutional team from ISB, Brigham & Women’s Hospital (BWH), the Centre National de la Recherche Scientifique (CNRS), and Institut Pasteur is interrogating the diverse capabilities of C. difficile by longitudinally profiling changes in its transcriptome, protein-DNA interactions, and phenotypes across dozens of environments and in vivo host-contexts. We are integrating these multi-omics datasets and generating systems-scale regulatory and metabolic network models to dissect conditionally active programs that control virulence (e.g., toxin production), colonization, and adaptive responses of C. difficile. These models and datasets will enable hypothesis-driven experimentation to characterize conditional vulnerabilities of C. difficile across diverse host-relevant contexts and enable rational formulation of effective therapy regimens.

