Metabolic Network model for C. difficile 630

 Metabolic model of C.difficile: Genome-scale metabolic models consists of curated biochemical reactions and can be used to investigate genotype to phenotype relationship. These genome-wide models have the potential to predict phenotypes in various environments.
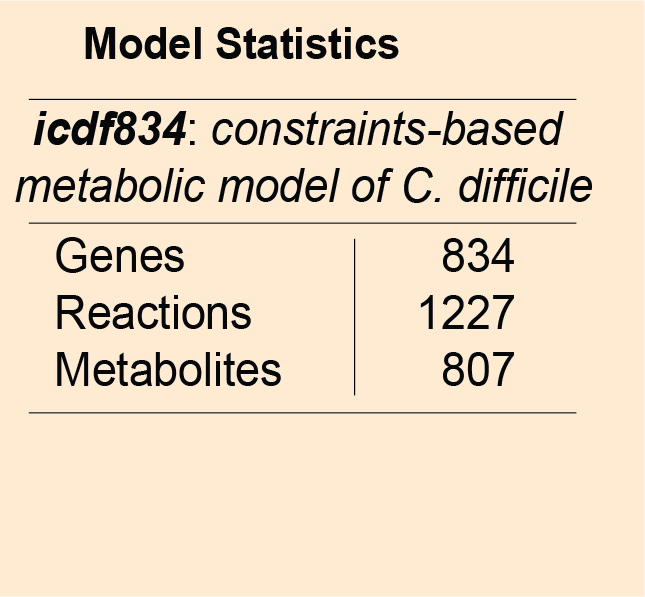
Metabolic model of C.difficile: Genome-scale metabolic models consists of curated biochemical reactions and can be used to investigate genotype to phenotype relationship. These genome-wide models have the potential to predict phenotypes in various environments. icdf834 is an updated metabolic network of C. difficile that builds on iMLTC806cdf and features 1227 reactions, 834 genes, and 807 metabolites. Genes in C. difficile are linked to the reactions based on gene-protein-reaction (GPR) associations. The performance of the icdf834 model is depicted in the form of a receiver operating characteristic (ROC) curve that shows the sensitivity and specificity of the model predictions for gene essentiality. The TnSeq analysis performed under nutrient-rich condition has been used to validate the predictions using the ROC curve.
Metabolic model adapted from: Kashaf, S., Angione, C. & Lió, P. Making life difficult for Clostridium difficile: augmenting the pathogen’s metabolic model with transcriptomic and codon usage data for better therapeutic target characterization. BMC Syst Biol 11, 25 (2017). https://doi.org/10.1186/s12918-017-0395-3
TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24. doi:10.1128/mBio.02383-14
Explore Metabolic Reactions
Displaying 111 - 120 of 1227| Desc | Reaction | GPR | Genes | Subsystem | ||
|---|---|---|---|---|---|---|
| Bio_CLP |
Peptidoglycan synthesis |
1.064 udpamagdapaa[c] + 1.064 udpaga[c] + 4.425 atp[c] + h2o[c] -> CLP[c] + 1.106 alaD[c] + 1.106 udp[c] + 4.425 adp[c] + 4.425 pi[c] | (CD630_26510) | CD630_26510 | Biomass Equation | Bookmark |
| Bio_prot |
Protein synthesis |
0.375 alaL[c] + 0.225 argL[c] + 0.441 asnL[c] + 0.399 aspL[c] + 0.087 cysL[c] + 0.167 glnL[c] + 0.517 gluL[c] + 0.442 gly[c] + 0.095 his[c] + 0.693 ileL[c] + 0.636 leuL[c] + 0.637 lysL[c] + 0.190 metL[c] + 0.304 pheL[c] + 0.192 proL[c] + 0.457 serL[c] + 0.344 thrL[c] + 0.043 trpL[c] + 0.286 tyrL[c] + 0.462 valL[c] + 39.94 atp[c] + h2o[c] -> Prot[c] + 39.94 adp[c] + 39.94 pi[c] | Biomass Equation | Bookmark | ||
| Bio_lip |
Lipid composition |
0.005699 phosglcditetdec[c] + 0.02308 phosglcdihexdec[c] + 0.007846 phosglcdioctdec[c] + 0.01447 dgludpalmgl[c] + 0.003517 dgludmygl[c] + 0.01119 mglucsyldpalmgl[c] + 0.00276 mglucsyldmygl[c] + 0.002845 palmphgl[c] + 0.0006934 myrphgl[c] + 0.0009777 strphglc[c] + 0.0005977 palmcdlpn[c] + 0.0001484 mycdlpn[c] + 0.0002022 strcdlpn[c] <=> Lip[c] | Biomass Equation | Bookmark | ||
| Bio_SPs |
Solute Pool |
0.0008 atp[c] + 0.1161 alaL[c] + 0.0288 argL[c] + 0.5959 aspL[c] + 0.5256 gluL[c] + 0.0086 glnL[c] + 0.0398 gly[c] + 0.0169 his[c] + 0.0481 ileL[c] + 0.0222 leuL[c] + 0.0405 lysL[c] + 0.0222 metL[c] + 0.0043 pheL[c] + 0.1293 proL[c] + 0.756 serL[c] + 0.0156 thrL[c] + 0.0017 thym[c] + 0.0162 valL[c] + 0.9947 pi[c] + 0.4775 glcAD[c] + 0.00033 acoa[c] + 0.0004 coa[c] + 0.0001 citrL[c] + 0.00067 fad[c] + 0.01433 nad[c] + 0.00033 nadh[c] + 0.00087 nadp[c] + 0.00267 nadph[c] -> SPs[c] | Biomass Equation | Bookmark | ||
| ID_525 |
acetyl-CoA:4-hydroxybutanoate CoA-transferase |
4hbut[c] + acoa[c] -> ac[c] + 4hcoa[c] | (CD630_23390) | CD630_23390 | Butanoate metabolism | Bookmark |
| ID_616 |
vinylacetyl-CoA delta3-delta2-isomerase |
vacoa[c] <=> ctncoa[c] | (CD630_23410) | CD630_23410 | Butanoate metabolism | Bookmark |
| ID_530 |
butanal:NADP+ oxidoreductase (CoA-acylating) |
btcoa[c] + nadph[c] + h[c] -> btal[c] + coa[c] + nadp[c] | (CD630_03340) or (CD630_29660) | CD630_03340, CD630_29660 | Butanoate metabolism | Bookmark |
| ID_519 |
(S)-3-hydroxybutanoyl-CoA:NADP+ oxidoreductase |
3hbcoa[c] + nadp[c] <=> aacoa[c] + nadph[c] + h[c] | (CD630_10580) | CD630_10580 | Butanoate metabolism | Bookmark |
| ID_72 |
butanoyl-CoA:acetate CoA-transferase |
btcoa[c] + ac[c] -> 2but[c] + acoa[c] | (CD630_26770 and CD630_26780) | CD630_26770, CD630_26780 | Butanoate metabolism | Bookmark |
| ID_502 |
(S)-acetoin:NAD+ oxidoreductase |
da[c] + nadh[c] + h[c] -> actnS[c] + nad[c] | (CD630_11820) | CD630_11820 | Butanoate metabolism | Bookmark |
