Metabolic Network model for C. difficile 630

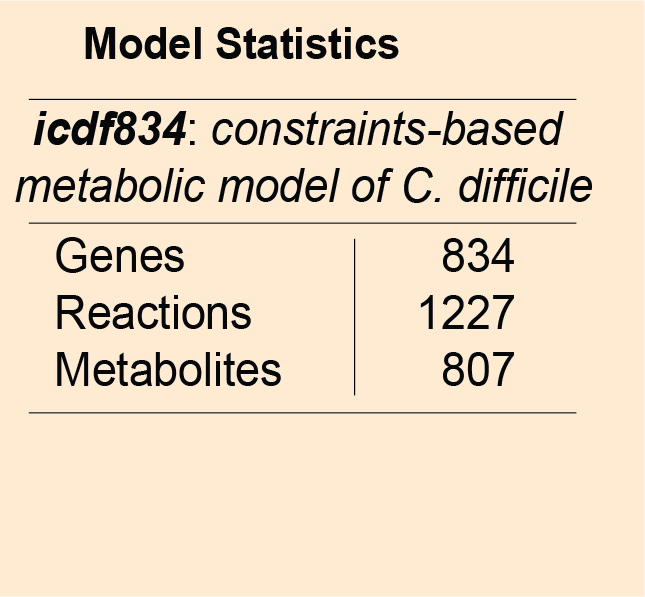
 Metabolic model of C.difficile: Genome-scale metabolic models consists of curated biochemical reactions and can be used to investigate genotype to phenotype relationship. These genome-wide models have the potential to predict phenotypes in various environments.
Metabolic model of C.difficile: Genome-scale metabolic models consists of curated biochemical reactions and can be used to investigate genotype to phenotype relationship. These genome-wide models have the potential to predict phenotypes in various environments. icdf834 is an updated metabolic network of C. difficile that builds on iMLTC806cdf and features 1227 reactions, 834 genes, and 807 metabolites. Genes in C. difficile are linked to the reactions based on gene-protein-reaction (GPR) associations. The performance of the icdf834 model is depicted in the form of a receiver operating characteristic (ROC) curve that shows the sensitivity and specificity of the model predictions for gene essentiality. The TnSeq analysis performed under nutrient-rich condition has been used to validate the predictions using the ROC curve.
Metabolic model adapted from: Kashaf, S., Angione, C. & Lió, P. Making life difficult for Clostridium difficile: augmenting the pathogen’s metabolic model with transcriptomic and codon usage data for better therapeutic target characterization. BMC Syst Biol 11, 25 (2017). https://doi.org/10.1186/s12918-017-0395-3
TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24. doi:10.1128/mBio.02383-14
Explore Metabolic Reactions
Displaying 311 - 320 of 1227| Desc | Reaction | GPR | Genes | Subsystem | ||
|---|---|---|---|---|---|---|
| ID_240 |
threonine synthase |
phser[c] + h2o[c] -> thrL[c] + pi[c] | (CD630_21180) | CD630_21180 | Glycine, serine and threonine metabolism | Bookmark |
| ID_130 |
betaine aldehyde:NAD+ oxidoreductase |
betald[c] + h2o[c] + nad[c] -> glyb[c] + nadh[c] + 2 h[c] | Glycine, serine and threonine metabolism | Bookmark | ||
| ID_536 |
L-cysteinylglycine dipeptidase |
cysgly[c] + h2o[c] -> cysL[c] + gly[c] | (CD630_07080) or (CD630_13000) | CD630_07080, CD630_13000 | Glycine, serine and threonine metabolism | Bookmark |
| ID_293 |
choline dehydrogenase (NADP+) |
chol[c] + nadp[c] -> betald[c] + nadph[c] + h[c] | (CD630_03340) or (CD630_30060) | CD630_03340, CD630_30060 | Glycine, serine and threonine metabolism | Bookmark |
| ID_330 |
L-threonine dehydratase |
thrL[c] -> 2obut[c] + nh3[c] | (CD630_25140) | CD630_25140 | Glycine, serine and threonine metabolism | Bookmark |
| ID_143 |
glycine hydroxymethyltransferase |
serL[c] + thf[c] <=> gly[c] + 510mlethf[c] + h2o[c] | (CD630_27260) or (CD630_28340) | CD630_27260, CD630_28340 | Glycine, serine and threonine metabolism | Bookmark |
| ID_452 |
phosphoglycerate dehydrogenase |
3pg[c] + nad[c] -> 3phpyr[c] + nadh[c] + h[c] | (CD630_09950) | CD630_09950 | Glycine, serine and threonine metabolism | Bookmark |
| ID_743 |
serine racemase |
serL[c] <=> serD[c] | (CD630_16280) or (CD630_28190) or (CD630_20280) or (CD630_04480) | CD630_16280, CD630_28190, CD630_20280, CD630_04480 | Glycine, serine and threonine metabolism | Bookmark |
| ID_665 |
3-phospho-L-serinephosphoserine transaminase |
3phpyr[c] + gluL[c] -> 3pser[c] + 2oglut[c] | (CD630_09940) | CD630_09940 | Glycine, serine and threonine metabolism | Bookmark |
| ID_132 |
homoserine dehydrogenase (NAD+) |
aspsald[c] + nadh[c] + h[c] -> hser[c] + nad[c] | (CD630_11660) or (CD630_15800) | CD630_11660, CD630_15800 | Glycine, serine and threonine metabolism | Bookmark |
