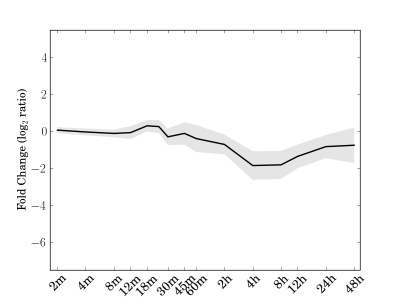
Module 24 Residual: 0.17
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0024 | v02 | 0.17 | -14.59 |
Displaying 1 - 21 of 21
| Cre01.g016500.t1.1 Dihydrolipoamide dehydrogenase | Cre02.g073700.t2.1 Uroporphyrinogen decarboxylase |
| Cre03.g210737.t1.1 | Cre04.g215000.t1.2 Beta-carotene ketolase |
| Cre06.g272450.t1.1 | Cre06.g278210.t1.1 phosphoglucomutase |
| Cre06.g287200.t2.1 | Cre07.g323600.t1.1 |
| Cre07.g356350.t1.1 Deoxyxylulose-5-phosphate synthase | Cre09.g405550.t1.2 Mitochondrial inner membrane translocase |
| Cre09.g416200.t1.2 high chlorophyll fluorescent 107 | Cre12.g491450.t1.1 |
| Cre12.g508000.t1.1 hydroxyproline-rich glycoprotein family protein | Cre12.g511900.t1.2 D-ribulose-5-phosphate-3-epimerase |
| Cre12.g513750.t1.1 Glutaredoxin family protein | Cre12.g519180.t2.1 elongation factor Ts family protein |
| Cre12.g546050.t1.2 1-deoxy-D-xylulose 5-phosphate reductoisomerase | Cre13.g569700.t1.1 |
| Cre13.g584850.t1.1 glutathione S-transferase PHI 10 | Cre17.g722900.t1.1 |
| Cre17.g739550.t1.1 Domain of unknown function (DUF1995) |
|
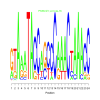
e.value: 3.4e-16 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: GTAaATACCAGCCCAAACTAAACC |
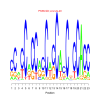
motif_0024_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 2.2e-20 Motif Bicluster: Width: 23 Number of Sites: 1 Consensus: cCacCaCCacCacCAcCnCCACC |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
| down-regulates |
Cre07.g341800.t1.2Submitted by admin on Tue, 05/19/2015 - 15:52nuclear factor Y, subunit B11 |
0.035214 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0070402 | NADPH binding |

Comments