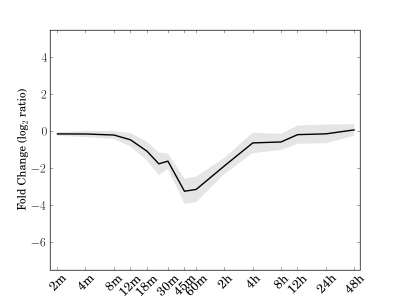
Module 68 Residual: 0.13
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0068 | v02 | 0.13 | -15.07 |
Displaying 1 - 15 of 15
| Cre01.g036600.t1.2 small RNA degrading nuclease 3 | Cre01.g042900.t1.2 Ribosomal RNA adenine dimethylase family protein |
| Cre02.g088700.t1.2 | Cre02.g092100.t1.2 |
| Cre02.g118300.t1.2 DEAD box RNA helicase (PRH75) | Cre03.g175650.t1.1 |
| Cre04.g217933.t1.1 | Cre04.g217947.t1.1 |
| Cre06.g304600.t1.2 Rhodanese/Cell cycle control phosphatase superfamily protein | Cre08.g358570.t1.1 Mitochondrial substrate carrier family protein |
| Cre08.g368500.t1.2 RNA-binding (RRM/RBD/RNP motifs) family protein | Cre09.g415750.t1.2 General substrate transporter, major facilitator superfamily |
| Cre12.g488050.t1.2 cell wall invertase 4 | Cre12.g505200.t1.2 DEA(D/H)-box RNA helicase family protein |
| Cre12.g558400.t1.1 |
|
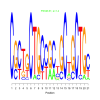
e.value: 1.4 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: CGCTgCTGCcGCtGCtGCTGc |
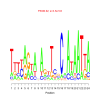
motif_0068_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 3500 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: TtttagtanAgtTcnCnAtAATtA |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0009451 | GO:0034470 | RNA modification | ncRNA processing |

Comments