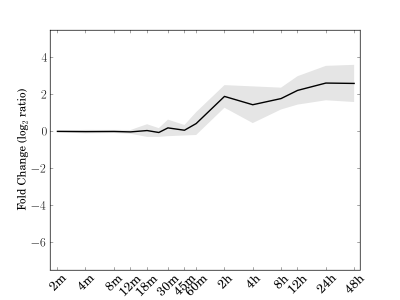
Module 82 Residual: 0.23
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0082 | v02 | 0.23 | -11.68 |
Displaying 1 - 20 of 20
| Cre02.g115450.t1.1 Adenylate/guanylate cyclase | Cre02.g142601.t1.1 |
| Cre03.g195500.t2.1 Acyl transferase/acyl hydrolase/lysophospholipase superfamily protein | Cre04.g211650.t1.2 |
| Cre04.g232802.t1.1 Protein phosphatase 2C family protein | Cre06.g258450.t2.1 |
| Cre06.g266200.t2.1 | Cre06.g278093.t1.1 exostosin family protein |
| Cre09.g417400.t2.1 origin of replication complex 1B | Cre12.g523000.t1.1 zinc finger protein 1 |
| Cre12.g537900.t1.1 | Cre13.g572750.t2.1 Copper amine oxidase family protein |
| Cre14.g617200.t1.1 RegA/RlsA-like protein | Cre14.g624800.t1.2 ARRESTIN DOMAIN CONTAINING PROTEIN |
| Cre14.g631400.t1.1 | Cre15.g636950.t1.2 |
| Cre16.g657050.t1.2 | Cre16.g659450.t1.2 alpha 1,4-glycosyltransferase family protein |
| Cre16.g685650.t1.1 glutamate receptor 3.6 | Cre17.g725550.t1.2 glucose-6-phosphate dehydrogenase 5 |
|
e.value: 0.000043 Motif Bicluster: Width: 19 Number of Sites: 1 Consensus: CgTgATAAcgTgtAcaTaT |
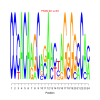
motif_0082_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 63 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: CCCACAaGCagAactAGgGGcCAc |
Displaying 1 - 3 of 3
| Interaction | Weight | |
|---|---|---|
| up-regulates |
Cre16.g673250.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51squamosa promoter binding protein-like 14, NRR1 |
0.056255 |
| up-regulates |
Cre10.g453500.t1.2Submitted by admin on Tue, 05/19/2015 - 15:51 |
0.060415 |
| down-regulates |
Cre12.g534450.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51SMAD/FHA domain-containing protein |
0.051983 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0004345 | glucose-6-phosphate dehydrogenase activi... |

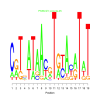
Comments