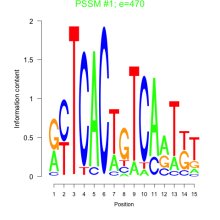
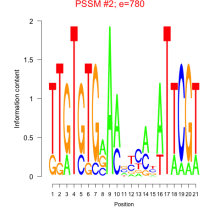
Phatr_bicluster_0002 Residual: 0.43
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0002 | 0.43 | Phaeodactylum tricornutum |
Displaying 1 - 78 of 78
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_13856 solute carrier family 25 (carnitine acylcarnitine translocase)member 20 (Mito_carr)
PHATRDRAFT_14607 pms2 postmeiotic segregation incre (mutl)
PHATRDRAFT_14735 qs (quinolinate synthase) quinolinate synthetase a (NadA superfamily)
PHATRDRAFT_15160 dynactin 25 kda subunit (LbetaH superfamily)
PHATRDRAFT_16991 uric acid-xanthine permease (Sulfate_transp superfamily)
PHATRDRAFT_21791 novel protein (ArfGap)
PHATRDRAFT_22554 alg2 protein (Glycosyltransferase_GTB_type superfamily)
PHATRDRAFT_22568 glycolate oxidase (FMN_dh)
PHATRDRAFT_23723 at3g05290 t12h1_26 (Mito_carr)
PHATRDRAFT_2394 cysteinyl-trna synthetase family protein (nt_trans superfamily)
PHATRDRAFT_2739 leucine-rich repeat transmembrane protein
PHATRDRAFT_2936 ubiquitin carboxyl-terminal hydrolase family protein (UCH)
PHATRDRAFT_30638 gpi-anchor transamidase precursor (Peptidase_C13 superfamily)
PHATRDRAFT_33251 conserved hypothetical
PHATRDRAFT_33624 matrix metalloproteinase (Peptidase_M11 superfamily)
PHATRDRAFT_37038 taurine catabolism dioxygenase (CAS_like superfamily)
PHATRDRAFT_37404 copia protein (gag-int-pol protein)
PHATRDRAFT_37459 probable hydrolase (HAD superfamily)
PHATRDRAFT_38848 xpg i-region familyexpressed (PIN_SF superfamily)
PHATRDRAFT_39081 pas pac sensor hybrid histidine kinase (HATPase_c)
PHATRDRAFT_39727 at4g32140 f10n7_50 (RhaT)
PHATRDRAFT_40023 core 1 udp-galactose:n-acetylgalactosamine-alpha-r beta- galactosy
PHATRDRAFT_42585 zinc finger in n-recognin family protein (zf-UBR)
PHATRDRAFT_43830 (Vps35 superfamily)
PHATRDRAFT_44021 potassium channel (Ion_trans_2)
PHATRDRAFT_46971 xpg-like endonuclease cg10670-pa (PIN_SF superfamily)
PHATRDRAFT_47583 short-chain dehydrogenase reductase sdr (SDR_c)
PHATRDRAFT_47798 flavin-containing monooxygenase
PHATRDRAFT_48289 dna helicase (DEXDc)
PHATRDRAFT_48393 2-oxoglutarate dehydrogenase e3 component (dihydrolipoamide dehydrogenase) (Lpd)
PHATRDRAFT_48525 zinc finger (zf-DHHC)
PHATRDRAFT_48645 pyridoxamine 5-phosphate oxidase-fmn-binding (Pyridox_oxidase)
PHATRDRAFT_48667 heat shock factor 6 (HSF_DNA-bind superfamily)
PHATRDRAFT_48752 (SfsA superfamily)
PHATRDRAFT_49198 (Sec7_N superfamily)
PHATRDRAFT_49902 asparagine synthase (glutamine-hydrolyzing) or related prot (Asn_Synthase_B_C)
PHATRDRAFT_49934 hyperpolarization-cyclic nucleotide-gated potassium channel 3
PHATRDRAFT_50274 dna polymerase epsilon subunit 2 isoform 1 (DNA_pol_E_B)
PHATRDRAFT_50401 glutamyl-trnaamidotransferase subunit a (GGCT_like superfamily)
PHATRDRAFT_51283 splicing factorsubunit 4 (RRM2_SF3B4)
PHATRDRAFT_52547 protein kinase (S_TKc)
PHATRDRAFT_7959 myb-like dna-binding domain containing protein (SANT)
PHATRDRAFT_8524 loc494751 protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0051336 | regulation of hydrolase activity | 0.000631938 | 1 | Phatr_bicluster_0002 |
| GO:0006423 | cysteinyl-tRNA aminoacylation | 0.0198767 | 1 | Phatr_bicluster_0002 |
| GO:0006281 | DNA repair | 0.0210699 | 1 | Phatr_bicluster_0002 |
| GO:0006529 | asparagine biosynthesis | 0.0264173 | 1 | Phatr_bicluster_0002 |
| GO:0006289 | nucleotide-excision repair | 0.0329159 | 1 | Phatr_bicluster_0002 |
| GO:0006298 | mismatch repair | 0.0393727 | 1 | Phatr_bicluster_0002 |
| GO:0043087 | regulation of GTPase activity | 0.0710389 | 1 | Phatr_bicluster_0002 |
| GO:0006813 | potassium ion transport | 0.125523 | 1 | Phatr_bicluster_0002 |
| GO:0006810 | transport | 0.132184 | 1 | Phatr_bicluster_0002 |
| GO:0006260 | DNA replication | 0.160121 | 1 | Phatr_bicluster_0002 |

Comments