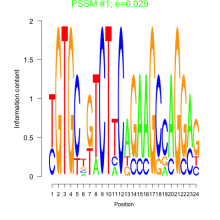
Phatr_bicluster_0020 Residual: 0.35
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0020 | 0.35 | Phaeodactylum tricornutum |
Displaying 1 - 38 of 38
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_16674 dna methyltransferase 2 (Cyt_C5_DNA_methylase superfamily)
PHATRDRAFT_16859 nad-dependent deacetylase sirtuin 2 homolog (SIR2 superfamily)
PHATRDRAFT_20188 elongation factor tu gtp binding domain containing 1 isoform 1 (Ras_like_GTPase superfamily)
PHATRDRAFT_29136 actin a (ACTIN)
PHATRDRAFT_33278 cytosolic 3-hydroxy-3-methylglutaryl-synthase (HMG_CoA_synt_N superfamily)
PHATRDRAFT_44031 riken cdnaisoform cra_c
PHATRDRAFT_44582 hsf2 protein (HSF_DNA-bind superfamily)
PHATRDRAFT_47369 (RecQ)
PHATRDRAFT_48375 guanylate cyclase (CHD)
PHATRDRAFT_48773 calmodulin 4 (calcium-binding protein dd112)
PHATRDRAFT_49225 msx-2 interacting nuclear target protein
PHATRDRAFT_49253 nadh dehydrogenase (CoA_CoA_reduc)
PHATRDRAFT_49373 d3-type cyclin
PHATRDRAFT_49948 cg9520-isoform b
PHATRDRAFT_50137 cog0579: dehydrogenase (DAO)
PHATRDRAFT_52468 p type cation copper-transporter (ZntA)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006342 | chromatin silencing | 0.0191218 | 1 | Phatr_bicluster_0020 |
| GO:0006259 | DNA metabolism | 0.0379055 | 1 | Phatr_bicluster_0020 |
| GO:0009190 | cyclic nucleotide biosynthesis | 0.0533044 | 1 | Phatr_bicluster_0020 |
| GO:0006306 | DNA methylation | 0.0624342 | 1 | Phatr_bicluster_0020 |
| GO:0000074 | regulation of cell cycle | 0.103975 | 1 | Phatr_bicluster_0020 |
| GO:0007242 | intracellular signaling cascade | 0.126943 | 1 | Phatr_bicluster_0020 |
| GO:0007165 | signal transduction | 0.141021 | 1 | Phatr_bicluster_0020 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.153654 | 1 | Phatr_bicluster_0020 |
| GO:0006118 | electron transport | 0.165064 | 1 | Phatr_bicluster_0020 |
| GO:0006464 | protein modification | 0.184674 | 1 | Phatr_bicluster_0020 |

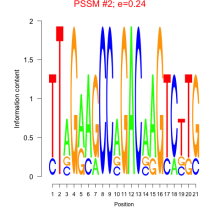
Comments