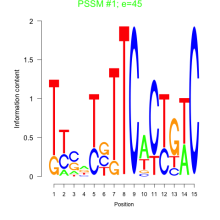
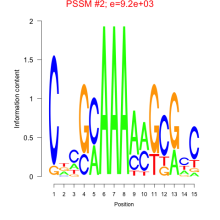
Phatr_bicluster_0226 Residual: 0.15
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0226 | 0.15 | Phaeodactylum tricornutum |
Displaying 1 - 17 of 17
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10544 (DUF1253)
PHATRDRAFT_14032 dead deah box (DEADc)
PHATRDRAFT_14068 chromosome 9 open reading frame 114 (Methyltrn_RNA_3)
PHATRDRAFT_1840 amidophosphoribosyltransferase (PRK09246)
PHATRDRAFT_22658 rna cyclase homolog (18S_RNA_Rcl1p)
PHATRDRAFT_23865 atp-dependent rna helicase-like protein (HA2)
PHATRDRAFT_41002 thif family (YgdL_like)
PHATRDRAFT_47420 mgc148433 protein (APH_ChoK_like superfamily)
PHATRDRAFT_47446 heat shock proteindomain protein (DnaJ)
PHATRDRAFT_47526 small nucleolar ribonucleoprotein complex component (Utp12 superfamily)
PHATRDRAFT_47947 glutamate-rich wd repeat containing 1 (COG2319)
PHATRDRAFT_48320 phytanoyl-dioxygenase family protein (PhyH superfamily)
PHATRDRAFT_50135 rio kinase 1 (RIO1_euk)
PHATRDRAFT_50246 (Utp14)
PHATRDRAFT_50326 superkiller viralicidic activity 2-like 2 (COG4581)
PHATRDRAFT_5128 proteasome component pre2 precursor (Ntn_hydrolase superfamily)
PHATRDRAFT_7346 mgc53444 protein (Ribosomal_L24e_L24)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009113 | purine base biosynthesis | 0.00296186 | 1 | Phatr_bicluster_0226 |
| GO:0009116 | nucleoside metabolism | 0.0132709 | 1 | Phatr_bicluster_0226 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.0648966 | 1 | Phatr_bicluster_0226 |
| GO:0006457 | protein folding | 0.161416 | 1 | Phatr_bicluster_0226 |
| GO:0006412 | protein biosynthesis | 0.216202 | 1 | Phatr_bicluster_0226 |
| GO:0008152 | metabolism | 0.364015 | 1 | Phatr_bicluster_0226 |

Comments