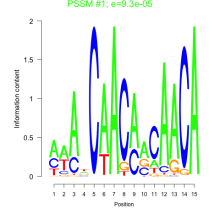
Thaps_bicluster_0009 Residual: 0.42
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0009 | 0.42 | Thalassiosira pseudonana |
Displaying 1 - 47 of 47
" class="views-fluidgrid-wrapper clear-block">
10046 hypothetical protein
10537 DEXDc
10768 hypothetical protein
11211 Reprolysin_5
11604 hypothetical protein
11629 hypothetical protein
12008 hypothetical protein
12079 hypothetical protein
12087 hypothetical protein
12153 hypothetical protein
1869 hypothetical protein
20703 hypothetical protein
23654 (Tp_AP2-EREBP6) regulator [Rayko]
23848 (Tp_HSF_3.5a) HSF_DNA-bind superfamily
23915 (Tp_Myb2R1) regulator [Rayko]
24602 hypothetical protein
24958 hypothetical protein
2509 hypothetical protein
2535 hypothetical protein
25483 hypothetical protein
25802 hypothetical protein
261895 GH18_chitinase-like superfamily
263284 (ZFP10) zinc finger protein similarity, PHD motif, DNA binding, transcription factor
263482 rad2
263734 LRR_RI superfamily
264838 (Tp_HSF_2.5e) regulator [Rayko]
2657 Abhydrolase_6
269452 hypothetical protein
3477 hypothetical protein
36631 UDPGlcNAc_PPase
4362 hypothetical protein
5511 hypothetical protein
5870 hypothetical protein
7245 hypothetical protein
7301 hypothetical protein
7359 (Tp_AP2-EREBP8) regulator [Rayko]
7394 (Tp_HSF_2.5i) HSF_DNA-bind superfamily
8054 hypothetical protein
8174 hypothetical protein
8420 UbiH
8799 hypothetical protein
ThpsCp008 (rbcS) ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit
ThpsCt004 (trnR(ucu))
ThpsCt008 (trnI(cau))
ThpsCt024 (trnR(acg))
ThpsCt026 (trnC(gca))
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006355 | regulation of transcription, DNA-dependent | 0.000289519 | 0.790097 | Thaps_bicluster_0009 |
| GO:0051336 | regulation of hydrolase activity | 0.0229404 | 1 | Thaps_bicluster_0009 |
| GO:0006725 | aromatic compound metabolism | 0.0704475 | 1 | Thaps_bicluster_0009 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.14198 | 1 | Thaps_bicluster_0009 |
| GO:0006281 | DNA repair | 0.14198 | 1 | Thaps_bicluster_0009 |
| GO:0008152 | metabolism | 0.146257 | 1 | Thaps_bicluster_0009 |
| GO:0005975 | carbohydrate metabolism | 0.221528 | 1 | Thaps_bicluster_0009 |
| GO:0006118 | electron transport | 0.596044 | 1 | Thaps_bicluster_0009 |
| GO:0006508 | proteolysis and peptidolysis | 0.626101 | 1 | Thaps_bicluster_0009 |


Comments