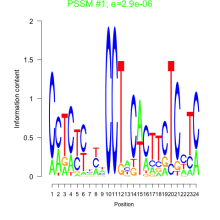
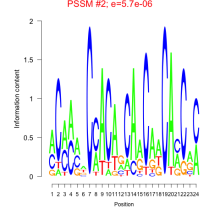
Thaps_bicluster_0065 Residual: 0.48
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0065 | 0.48 | Thalassiosira pseudonana |
Displaying 1 - 39 of 39
" class="views-fluidgrid-wrapper clear-block">
10063 Pep_deformylase superfamily
10149 hypothetical protein
10249 hypothetical protein
10928 hypothetical protein
11315 CAP10 superfamily
12116 (Tp_HSF_3.5b) HSF_DNA-bind superfamily
17946 Lactamase_B superfamily
20349 GH38-57_N_LamB_YdjC_SF superfamily
21312 hypothetical protein
2173 hypothetical protein
22032 2OG-FeII_Oxy_5 superfamily
22592 CBS_pair_10
23338 hypothetical protein
23623 hypothetical protein
24031 hypothetical protein
24320 hypothetical protein
24562 hypothetical protein
25389 hypothetical protein
25543 hypothetical protein
25871 hypothetical protein
25872 hypothetical protein
260925 nucleoside_deaminase
261078 Na_H_Exchanger
262679 MgtA
262877 Peptidase_C69 superfamily
264694 ACD_sHsps-like
269683 NBD_sugar-kinase_HSP70_actin superfamily
35129 (Tp_Myb2R9) PLN03142
36787 HMGB-UBF_HMG-box
36970 Fe-S_biosyn superfamily
4202 (Tp_bHLH8_PAS) pas-domain protein
5653 hypothetical protein
7372 Tryp_SPc
7786 hypothetical protein
7924 hypothetical protein
8216 hypothetical protein
9840 MntH superfamily
985 hypothetical protein
9963 ATS1
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015992 | proton transport | 0.0396209 | 1 | Thaps_bicluster_0065 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.0450164 | 1 | Thaps_bicluster_0065 |
| GO:0006813 | potassium ion transport | 0.0576102 | 1 | Thaps_bicluster_0065 |
| GO:0007165 | signal transduction | 0.10245 | 1 | Thaps_bicluster_0065 |
| GO:0006812 | cation transport | 0.109729 | 1 | Thaps_bicluster_0065 |
| GO:0009058 | biosynthesis | 0.138303 | 1 | Thaps_bicluster_0065 |
| GO:0006508 | proteolysis and peptidolysis | 0.179105 | 1 | Thaps_bicluster_0065 |
| GO:0006412 | protein biosynthesis | 0.365269 | 1 | Thaps_bicluster_0065 |
| GO:0006810 | transport | 0.396381 | 1 | Thaps_bicluster_0065 |
| GO:0008152 | metabolism | 0.681496 | 1 | Thaps_bicluster_0065 |

Comments