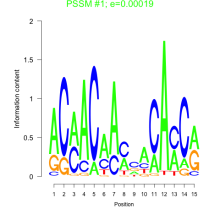
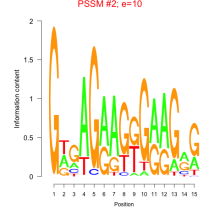
Thaps_bicluster_0104 Residual: 0.31
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0104 | 0.31 | Thalassiosira pseudonana |
Displaying 1 - 24 of 24
" class="views-fluidgrid-wrapper clear-block">
10088 hypothetical protein
10414 FAD_binding_7 superfamily
12037 hypothetical protein
12234 Carboxyl_trans
20563 hypothetical protein
21604 Abhydrolase_3
22650 17beta-HSD1_like_SDR_c
25891 Lipase_3
263937 Glyco_hydrolase_16 superfamily
264285 crotonase-like
264782 DUF4281
268208 HAD_2
2936 LPLAT_AGPAT-like
3258 PSII_Pbs27
3379 hypothetical protein
3435 Rotamase
35005 crypto_DASH
4737 hypothetical protein
5715 AhpC-TSA_2
5945 AarF
7512 hypothetical protein
7740 hypothetical protein
8014 hypothetical protein
907 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008152 | metabolism | 0.000726612 | 1 | Thaps_bicluster_0104 |
| GO:0006281 | DNA repair | 0.00379335 | 1 | Thaps_bicluster_0104 |
| GO:0006629 | lipid metabolism | 0.0584155 | 1 | Thaps_bicluster_0104 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.111884 | 1 | Thaps_bicluster_0104 |
| GO:0006468 | protein amino acid phosphorylation | 0.362866 | 1 | Thaps_bicluster_0104 |

Comments