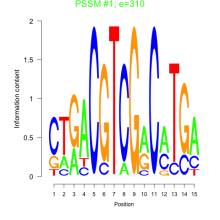
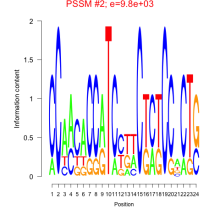
Thaps_bicluster_0159 Residual: 0.26
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0159 | 0.26 | Thalassiosira pseudonana |
Displaying 1 - 20 of 20
" class="views-fluidgrid-wrapper clear-block">
10360 (Tp_bZIP24a) regulator [Rayko]
22208 COG0075
22396 hypothetical protein
233 CdCA1
260953 COG2423
263924 MFS
264181 hypothetical protein
269696 hypothetical protein
269942 SHMT
28521 (GDCH) GCS_H
34125 CdCA1
36208 (GDCT) PLN02319
3974 (UroD2) URO-D
39799 (GDCP) PLN02414
412 (ALAT_1) PTZ00377
4819 Bestrophin superfamily
4820 Bestrophin superfamily
6528 hypothetical protein
6529 hypothetical protein
9903 (Tp_bZIP24b) regulator [Rayko]
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006544 | glycine metabolism | 0.000023 | 0.0634592 | Thaps_bicluster_0159 |
| GO:0006546 | glycine catabolism | 0.0000383 | 0.105534 | Thaps_bicluster_0159 |
| GO:0006563 | L-serine metabolism | 0.00619065 | 1 | Thaps_bicluster_0159 |
| GO:0006779 | porphyrin biosynthesis | 0.0225308 | 1 | Thaps_bicluster_0159 |
| GO:0009058 | biosynthesis | 0.108157 | 1 | Thaps_bicluster_0159 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.129013 | 1 | Thaps_bicluster_0159 |
| GO:0006810 | transport | 0.321719 | 1 | Thaps_bicluster_0159 |

Comments