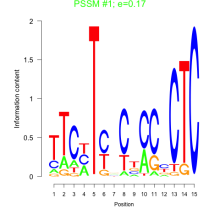
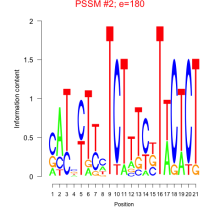
Thaps_bicluster_0187 Residual: 0.36
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0187 | 0.36 | Thalassiosira pseudonana |
Displaying 1 - 30 of 30
" class="views-fluidgrid-wrapper clear-block">
10191 SUL1
10465 hypothetical protein
10626 hypothetical protein
1081 hypothetical protein
12570 DEXDc
20907 hypothetical protein
20925 hypothetical protein
21852 hypothetical protein
22124 (Tp_HSF_1.2a) HSF_DNA-bind
22764 hypothetical protein
23497 hypothetical protein
23754 hypothetical protein
24827 CHD
24955 hypothetical protein
25397 MviM
25438 (Tp_fungal_TRF1b) regulator [Rayko]
25873 hypothetical protein
260818 DUF1126 superfamily
261639 (Helicase_1) rad3
262517 Glyco_18
35531 SET
4047 hypothetical protein
4050 hypothetical protein
5829 hypothetical protein
6186 hypothetical protein
6883 Peptidase_C19 superfamily
8408 hypothetical protein
9131 SCP superfamily
9790 hypothetical protein
991 PROCN superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006289 | nucleotide-excision repair | 0.0154159 | 1 | Thaps_bicluster_0187 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 0.0455847 | 1 | Thaps_bicluster_0187 |
| GO:0009190 | cyclic nucleotide biosynthesis | 0.066187 | 1 | Thaps_bicluster_0187 |
| GO:0006096 | glycolysis | 0.108945 | 1 | Thaps_bicluster_0187 |
| GO:0007242 | intracellular signaling cascade | 0.122787 | 1 | Thaps_bicluster_0187 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.147198 | 1 | Thaps_bicluster_0187 |
| GO:0000074 | regulation of cell cycle | 0.170973 | 1 | Thaps_bicluster_0187 |
| GO:0005975 | carbohydrate metabolism | 0.209229 | 1 | Thaps_bicluster_0187 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.245811 | 1 | Thaps_bicluster_0187 |
| GO:0008152 | metabolism | 0.363828 | 1 | Thaps_bicluster_0187 |

Comments