Regulatory Modules
Displaying 801 - 810 of 1058Desulfovibrio vulgaris Hildenborough
| Title | Motif 1 | Motif 2 | Residual | Bicluster Genes | Selected GO Terms |
|---|---|---|---|---|---|
| bicluster_0149 |
4000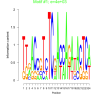 |
79000 |
0.35 | DVU2595, DVU2664, DVU0545, DVU1433, DVU1696, DVU1884, DVU2165, DVU2327, DVU2426, DVU2480, DVU2820 | response to stimulus |
| bicluster_0151 |
180 |
3200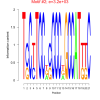 |
0.44 | DVU0873, DVU0874, DVU0956, DVU1287, DVU1288, DVU1289, DVU1293, DVU2519, DVU2922, DVU2925, DVU2927, DVU2928, DVU2929, DVU1574, DVU2231, DVU2518, DVU2923, DVU2924, DVU2926 | transcription, DNA-templated, RNA biosynthetic process, gene expression, RNA metabolic process, organelle |
| bicluster_0152 |
640 |
3800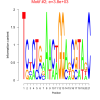 |
0.43 | DVU1486, DVU1494, DVU1495, DVU1497, DVU1501, DVU1503, DVU1523, DVU1487, DVU1489, DVU1502, DVU1504, DVU1505, DVU1506, DVU1972 | hydrolase activity, endonuclease activity, nuclease activity, peptidase activity, hydrolase activity, acting on ester bond... |
| bicluster_0154 |
620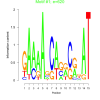 |
18000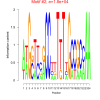 |
0.38 | DVU0670, DVU0967, DVU1276, DVU1626, DVU1662, DVU1671, DVU1672, DVU2356, DVU2367, DVU1027, DVU1294, DVU1919, DVU2306, DVU2842, DVU2891 | DNA (cytosine-5-)-methyltransferase acti..., inorganic phosphate transmembrane transp..., O-acyltransferase activity, DNA-methyltransferase activity, phosphate transmembrane transporter acti... |
| bicluster_0158 |
2700 |
2900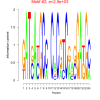 |
0.38 | DVU0857, DVU0865, DVU0994, DVU1267, DVU1602, DVU2470, DVU0242, DVU0758, DVU1278, DVU1336, DVU1337, DVU1874, DVU2310 | ATP binding, adenyl ribonucleotide binding, adenyl nucleotide binding, purine ribonucleoside triphosphate bindi..., nucleoside binding |
| bicluster_0159 |
70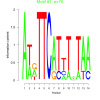 |
44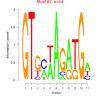 |
0.34 | DVU1510, DVU1692, DVU2179, DVU2220, DVU2686, DVU2836, DVU2839, DVUA0017, DVU1509, DVU1519 | DNA binding, nucleic acid binding, sequence-specific DNA binding |
| bicluster_0163 |
1300 |
21000 |
0.40 | DVU0506, DVU1246, DVU1292, DVU1682, DVU1843, DVU2371, DVU3070, DVUA0006, DVUA0068, DVUA0076, DVU2051, DVU2372, DVU3088 | DNA recombination, DNA metabolic process, hydrolase activity, acting on carbon-nit..., hydrolase activity, acting on carbon-nit... |
| bicluster_0164 |
2300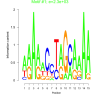 |
4000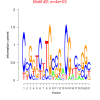 |
0.38 | DVU0074, DVU0657, DVU2227, DVU2323, DVU3174, DVU0153, DVU0157, DVU0321, DVU0792, DVU0793, DVU1041, DVU1242, DVU1407, DVU3186 | TAT protein transport complex, protein complex, racemase and epimerase activity, racemase and epimerase activity, acting ..., cyclic nucleotide binding |
| bicluster_0165 |
10000 |
1500 |
0.42 | DVU1511, DVU2155, DVU2173, DVU2540, DVU3306, DVU1518, DVU1565, DVU1752, DVU2185, DVU2189, DVU2253, DVU3044, DVU3260, DVU3285, DVU3329 | |
| bicluster_0167 |
0.00011 |
0.044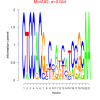 |
0.49 | DVU0131, DVU0132, DVU0133, DVU0587, DVU0684, DVU1382, DVU1838, DVU1943, DVU2044, DVU2489, DVU3344, DVUA0029, DVUA0127, DVU0254, DVU0401, DVU1383, DVU1722, DVU1839, DVU2469, DVU2486, DVU3346 | cell redox homeostasis, cellular homeostasis, homeostatic process, regulation of biological quality, N-acetyltransferase activity |

