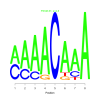
Module 126 Residual: 0.51
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0126 | v02 | 0.51 | -6.66 |
| Rv0072 Glutamine-transport transmembrane protein ABC transporter | Rv0073 Glutamine-transport ATP-binding protein ABC transporter |
| Rv0346c L-asparagine permease | Rv0433 Carboxylate-amine ligase |
| Rv0497 Possible membrane protein | Rv0719 LSU ribosomal protein L6p (L9e) |
| Rv1436 NAD-dependent glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12) | Rv1437 Phosphoglycerate kinase (EC 2.7.2.3) |
| Rv1438 Triosephosphate isomerase (EC 5.3.1.1) | Rv1980c Immunogenic protein MPT64/MPB64 precursor |
| Rv3920c RNA-binding protein Jag | Rv3921c Inner membrane protein translocase component YidC, long form |
| Rv0431 Tuberculin related peptide | Rv1241 |
| Rv2953 Trans-acting enoyl reductase | Rv2956 gsc |
| Rv3646c DNA topoisomerase I (EC 5.99.1.2) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006096 | glycolysis |
|
GO:0005886 | plasma membrane |
|
GO:0005887 | integral to plasma membrane |
|
GO:0009405 | pathogenesis |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0006412 | translation |
|
GO:0005622 | intracellular |
|
GO:0005840 | ribosome |
|
GO:0003735 | structural constituent of ribosome |
|
GO:0005618 | cell wall |
|
GO:0040007 | growth |
|
GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity |
|
GO:0005576 | extracellular region |
|
GO:0005829 | cytosol |
|
GO:0004618 | phosphoglycerate kinase activity |
|
GO:0004807 | triose-phosphate isomerase activity |
|
GO:0042803 | protein homodimerization activity |
|
GO:0009267 | cellular response to starvation |
|
GO:0045927 | positive regulation of growth |
|
GO:0009247 | glycolipid biosynthetic process |
|
GO:0071766 | Actinobacterium-type cell wall biogenesis |
|
GO:0003917 | DNA topoisomerase type I activity |
|
GO:0000287 | magnesium ion binding |



















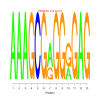
Discussion