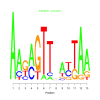
Module 167 Residual: 0.51
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0167 | v02 | 0.51 | -8.58 |
| Rv0158 Transcriptional regulator, TetR family | Rv0330c Transcriptional regulator, TetR family |
| Rv0474 \Transcriptional regulator, XRE family\\\"" | Rv0475 Iron-regulated heparin binding hemagglutinin HbhA (Adhesin) |
| Rv0711 Arylsulfatase (EC 3.1.6.1) | Rv0735 RNA polymerase sigma-70 factor |
| Rv0736 Possible membrane protein | Rv1169c (MTV005.05c), len: 100. similar to the N-terminal region of the M. tuberculosis PE family of proteinseg. TR:O05297 (EMBL:Z93777) MTCI364.07 (99 aa), fasta scores; opt: 209 z-score: 407.6 E(): 1.6e-15, 37.4% identityin 99 aa overlap. Also simlar to the N-terminus of M. tuberculosis TR:P77909 (EMBL:U76006) ESTERASE/LIPASE (EC 3.1.1.3) (TRIACYLGLYCEROL LIPASE) (437 aa), fasta scores; opt: 193z-score: 381.8 E(): 4.4e-14, 37.2% identity in 94 aa overlap. Contains a helix-turn-heix motiffrom aa 88-109(+2.76 SD) |
| Rv1532c Putative uncharacterized protein BCG_1584c | Rv3863 Probable forkhead-associated protein |
| Rv0068 Oxidoreductase/Short-chain dehydrogenase | Rv0712 Sulfatase modifying factor 1 precursor (C-alpha-formyglycine- generating enzyme 1) |
| Rv3061c Butyryl-CoA dehydrogenase (EC 1.3.99.2) | Rv3249c Transcriptional regulator, TetR family |
| Rv3250c Rubredoxin | Rv3251c Rubredoxin |
| Rv3252c Alkane-1 monooxygenase (EC 1.14.15.3) |
|
GO:0005829 | cytosol |
|
GO:0005515 | protein binding |
|
GO:0005618 | cell wall |
|
GO:0005886 | plasma membrane |
|
GO:0007157 | heterophilic cell-cell adhesion |
|
GO:0008201 | heparin binding |
|
GO:0009986 | cell surface |
|
GO:0042803 | protein homodimerization activity |
|
GO:0044078 | positive regulation by symbiont of host receptor-mediated endocytosis |
|
GO:0010033 | response to organic substance |
|
GO:0006355 | regulation of transcription, DNA-dependent |
|
GO:0040007 | growth |
|
GO:0004806 | triglyceride lipase activity |
|
GO:0052572 | response to host immune response |
|
GO:0018083 | peptidyl-L-3-oxoalanine biosynthetic process from peptidyl-cysteine or peptidyl-serine |
|
GO:0018685 | alkane 1-monooxygenase activity |
|
GO:0080133 | midchain alkane hydroxylase activity |



















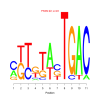
Discussion