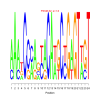
Module 176 Residual: 0.50
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0176 | v02 | 0.50 | -23.71 |
| Rv1095 Predicted ATPase related to phosphate starvation-inducible protein PhoH | Rv1126c |
| Rv1127c Pyruvate,phosphate dikinase (EC 2.7.9.1) | Rv2895c Iron utilization protein |
| Rv3688c Transamidase GatB domain protein | Rv3757c L-proline glycine betaine ABC transport system permease protein ProW (TC 3.A.1.12.1) |
| Rv3758c L-proline glycine betaine ABC transport system permease protein ProV (TC 3.A.1.12.1) | Rv3760 Cell division protein DivIC (FtsB), stabilizes FtsL against RasP cleavage |
| Rv3766 | Rv1104 Carboxylesterase (EC 3.1.1.1) |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0050242 | pyruvate, phosphate dikinase activity |
|
GO:0010106 | cellular response to iron ion starvation |
|
GO:0015891 | siderophore transport |
|
GO:0005215 | transporter activity |
|
GO:0006810 | transport |
|
GO:0016020 | membrane |
|
GO:0005488 | binding |
|
GO:0031460 | glycine betaine transport |
|
GO:0075136 | response to host |
|
GO:0005524 | ATP binding |
|
GO:0016887 | ATPase activity |












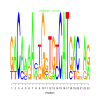
Discussion