Module 287 Residual: 0.56
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0287 | v02 | 0.56 | -12.32 |
| Rv0908 Cation-transporting ATPase, E1-E2 family | Rv1023 Enolase (EC 4.2.1.11) |
| Rv1024 Cell division protein DivIC (FtsB), stabilizes FtsL against RasP cleavage | Rv1025 FIG004853: possible toxin to DivIC |
| Rv1026 Exopolyphosphatase (EC 3.6.1.11) | Rv1281c Oligopeptide transport system permease protein OppD (TC 3.A.1.5.1) |
| Rv1282c Oligopeptide transport system permease protein OppC (TC 3.A.1.5.1) | Rv1283c Oligopeptide transport system permease protein OppB (TC 3.A.1.5.1) |
| Rv1522c Putative membrane protein MmpL12 | Rv0069c L-serine dehydratase (EC 4.3.1.17) |
| Rv0070c Serine hydroxymethyltransferase (EC 2.1.2.1) | Rv0103c Lead, cadmium, zinc and mercury transporting ATPase (EC 3.6.3.3) (EC 3.6.3.5); Copper-translocating P-type ATPase (EC 3.6.3.4) |
| Rv1609 Anthranilate synthase, aminase component (EC 4.1.3.27) | Rv1610 |
| Rv3392c Cyclopropane-fatty-acyl-phospholipid synthase (EC 2.1.1.79) | Rv3393 Inosine-uridine preferring nucleoside hydrolase (EC 3.2.2.1) |
|
GO:0004634 | phosphopyruvate hydratase activity |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0004309 | exopolyphosphatase activity |
|
GO:0005215 | transporter activity |
|
GO:0016020 | membrane |
|
GO:0006810 | transport |
|
GO:0005829 | cytosol |
|
GO:0052572 | response to host immune response |
|
GO:0004372 | glycine hydroxymethyltransferase activity |
|
GO:0004049 | anthranilate synthase activity |
|
GO:0008825 | cyclopropane-fatty-acyl-phospholipid synthase activity |
|
GO:0046500 | S-adenosylmethionine metabolic process |
|
GO:0071766 | Actinobacterium-type cell wall biogenesis |
|
GO:0071768 | mycolic acid biosynthetic process |


















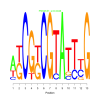

Discussion