Module 535 Residual: 0.57
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0535 | v02 | 0.57 | -7.49 |
| Rv3109 | Rv3110 Pterin-4-alpha-carbinolamine dehydratase (EC 4.2.1.96) |
| Rv3111 Molybdenum cofactor biosynthesis protein MoaC | Rv3112 Molybdenum cofactor biosynthesis protein MoaD |
| Rv3309c Xanthine-guanine phosphoribosyltransferase (EC 2.4.2.22) | Rv0187 O-methyltransferase |
| Rv0972c Acyl-CoA dehydrogenase, short-chain specific (EC 1.3.99.2) | Rv1050 Oxidoreductase, short-chain dehydrogenase/reductase family (EC 1.1.1.-) |
| Rv1263 Amidase (EC 3.5.1.4) | Rv1266c Serine/threonine protein kinase (EC 2.7.11.1) |
| Rv1514c Putative glycosyl transferase | Rv1719 Transcriptional regulator, IclR family |
| Rv1755c phospholipase C | Rv1802 PPE family protein |
| Rv1890c | Rv2742c |
| Rv3313c Adenosine deaminase (EC 3.5.4.4) | Rv3525c carbonic anhydrase, family 3 |
| Rv3654c |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0006777 | Mo-molybdopterin cofactor biosynthetic process |
|
GO:0005576 | extracellular region |
|
GO:0006790 | sulfur compound metabolic process |
|
GO:0004845 | uracil phosphoribosyltransferase activity |
|
GO:0008470 | isovaleryl-CoA dehydrogenase activity |
|
GO:0003995 | acyl-CoA dehydrogenase activity |
|
GO:0055114 | oxidation-reduction process |
|
GO:0004672 | protein kinase activity |
|
GO:0004674 | protein serine/threonine kinase activity |
|
GO:0005887 | integral to plasma membrane |
|
GO:0006950 | response to stress |
|
GO:0043085 | positive regulation of catalytic activity |
|
GO:0043086 | negative regulation of catalytic activity |
|
GO:0043388 | positive regulation of DNA binding |
|
GO:0045926 | negative regulation of growth |
|
GO:0045893 | positive regulation of transcription, DNA-dependent |
|
GO:0046777 | protein autophosphorylation |
|
GO:0046890 | regulation of lipid biosynthetic process |
|
GO:0052572 | response to host immune response |
|
GO:0034480 | phosphatidylcholine phospholipase C activity |
|
GO:0005829 | cytosol |
|
GO:0004000 | adenosine deaminase activity |





















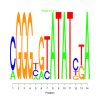
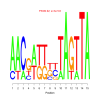
Discussion