Module 275
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.33 | 23 | 61 | NA | NA |
Bicluster expression profile
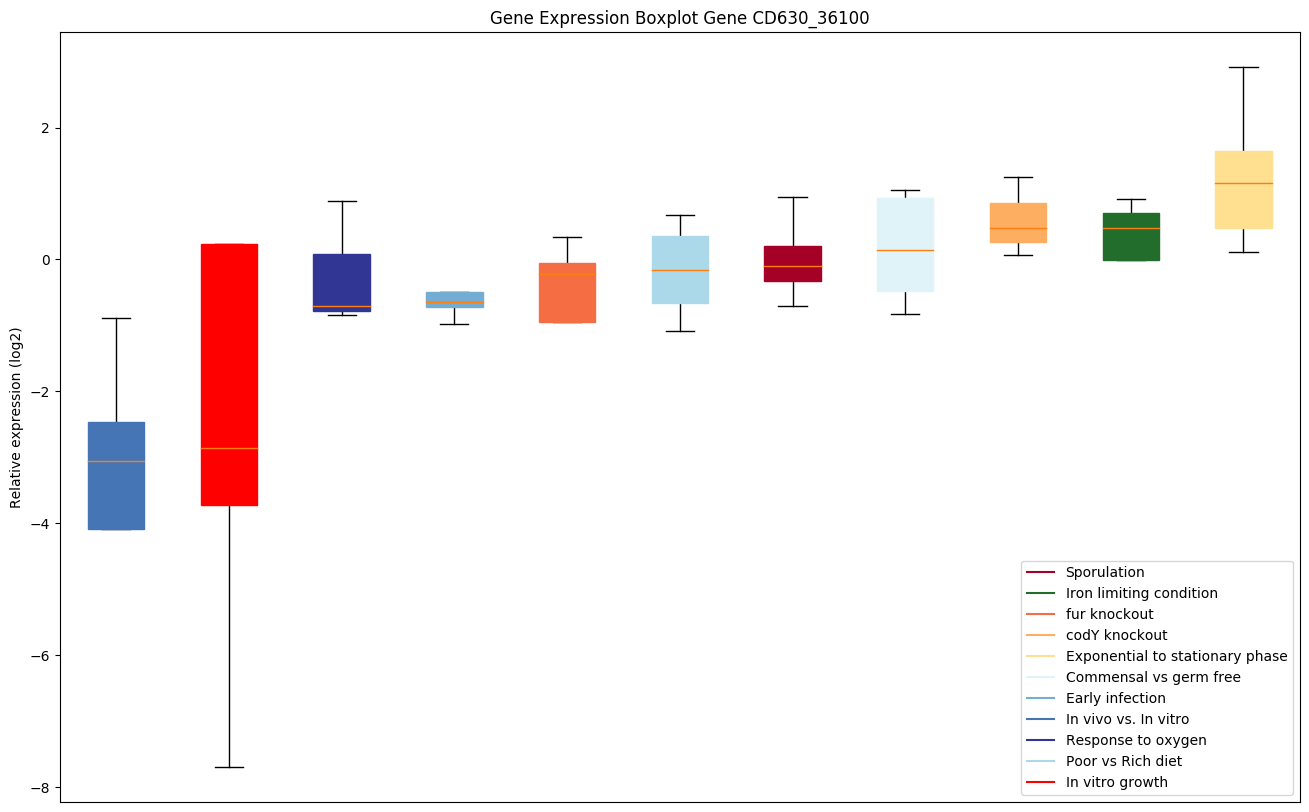
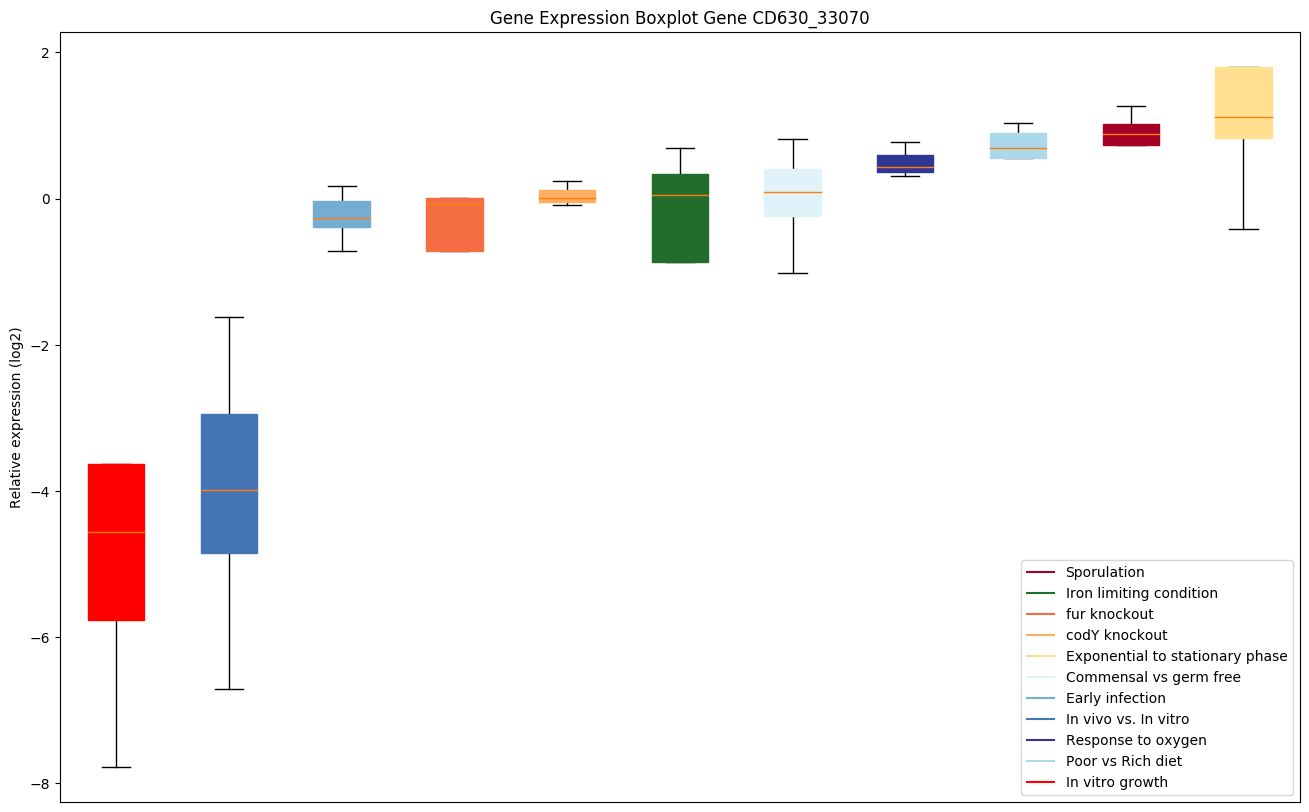
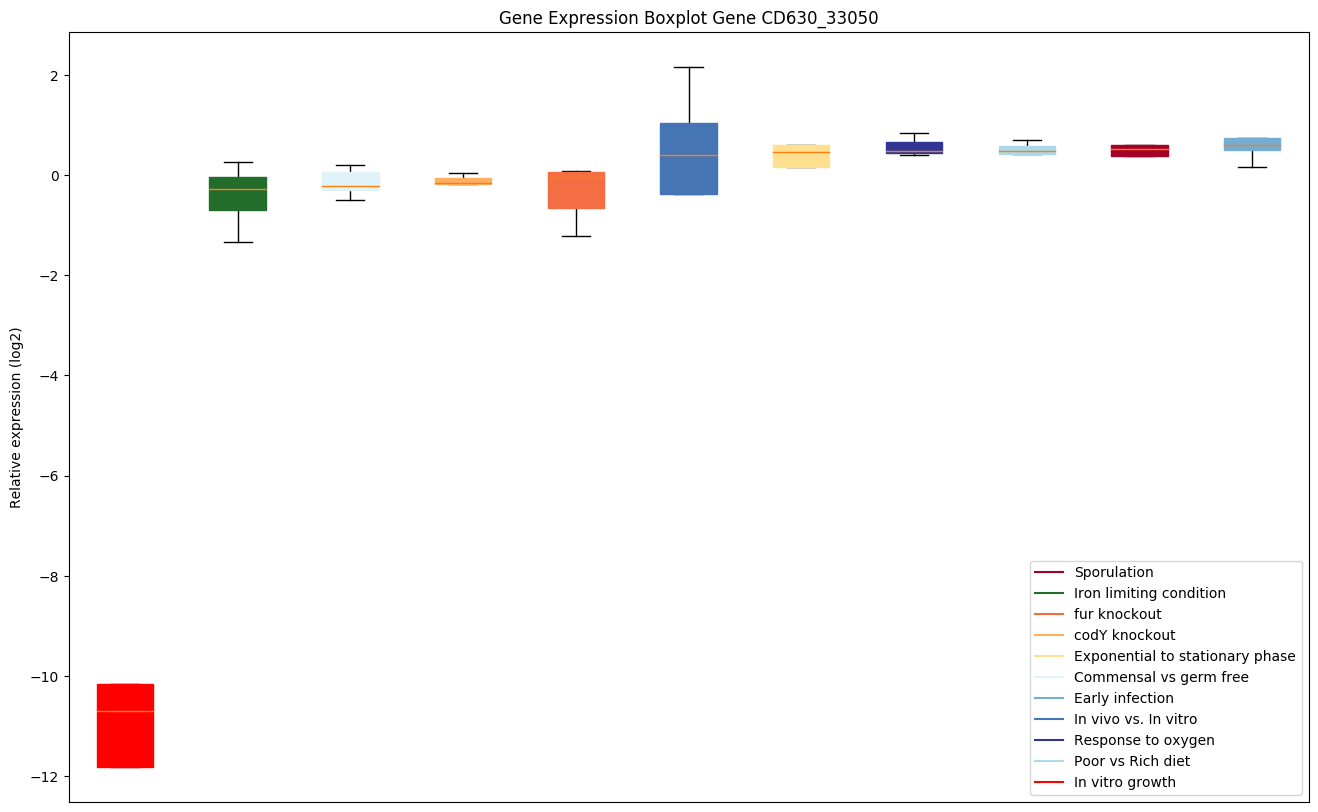
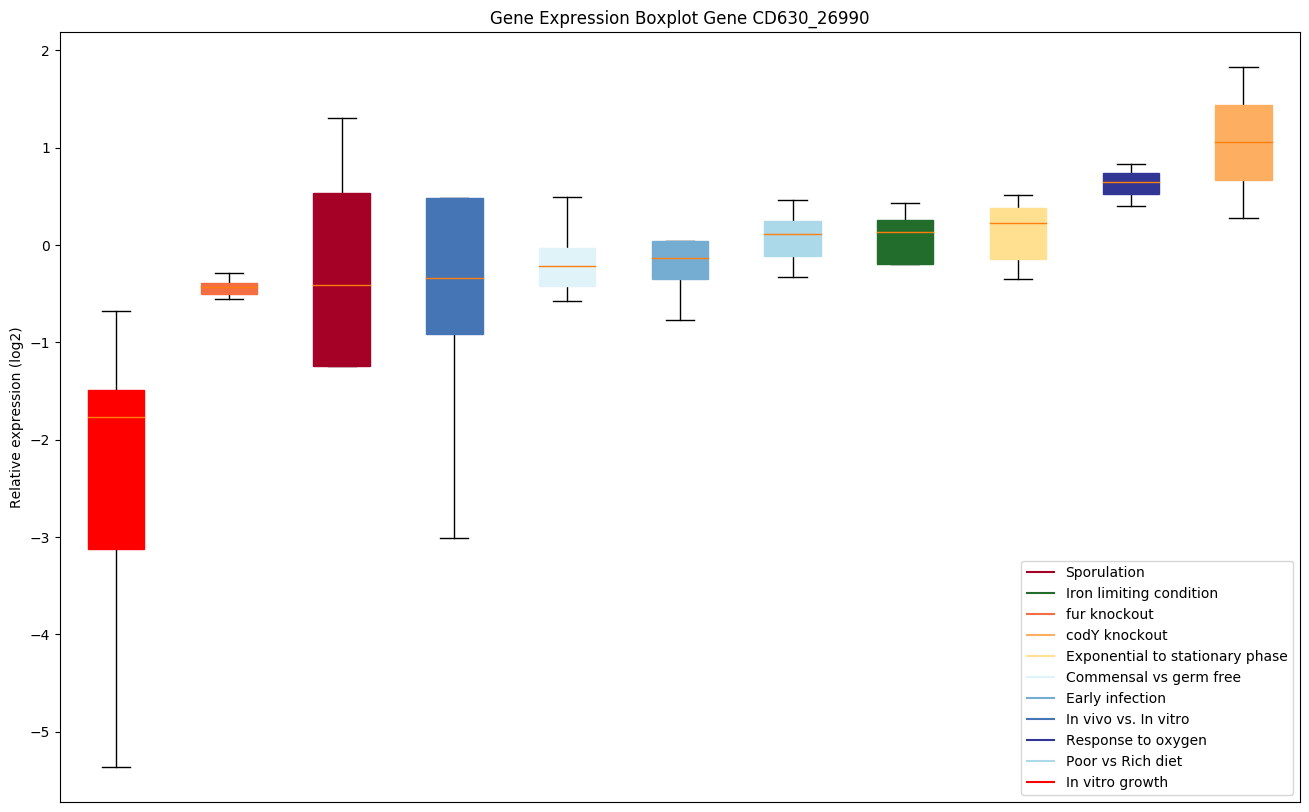
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
| TF | Module | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
|
ccpA Transcriptional regulator, LacI family |
275 | 6 | 0.00 | 0.03 |
The module is significantly enriched with genes associated to the indicated functional terms
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
| CD630_27470 | Putative ubiquinone biosynthesis protein |
 |
|||||
| CD630_33040 | clpX | ATP-dependent Clp protease ATP-binding subunitclpX | ATP-dependent specificity component of the Clp protease. It directs the protease to specific substrates. Can perform chaperone functions in the absence of ClpP. | Yes |
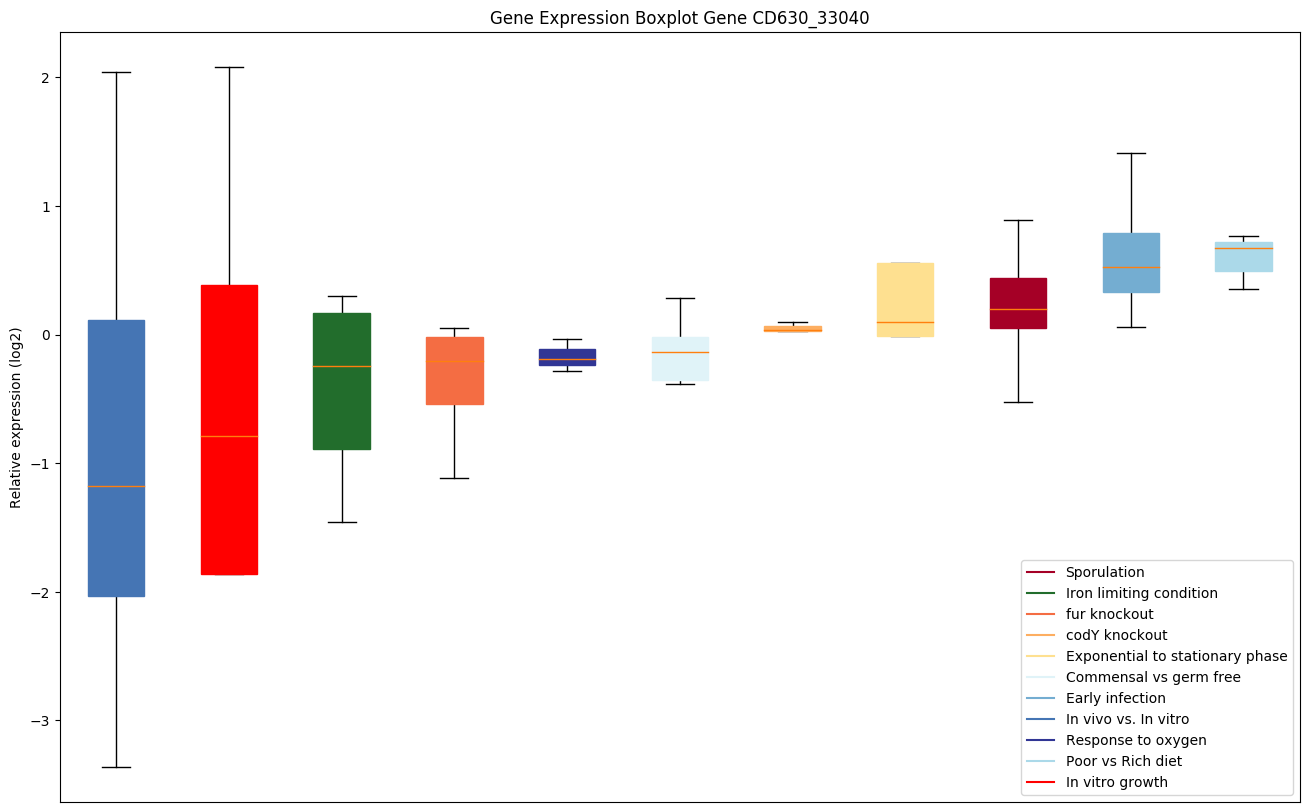 |
||
| CD630_35590 | ftsH2 | Cell division protease FtsH2 | Acts as a processive, ATP-dependent zinc metallopeptidase for both cytoplasmic and membrane proteins. Plays a role in the quality control of integral membrane proteins. |
 |
|||
| CD630_26980 | Putative membrane protein |
 |
|||||
| CD630_36090 | Conserved hypothetical protein |
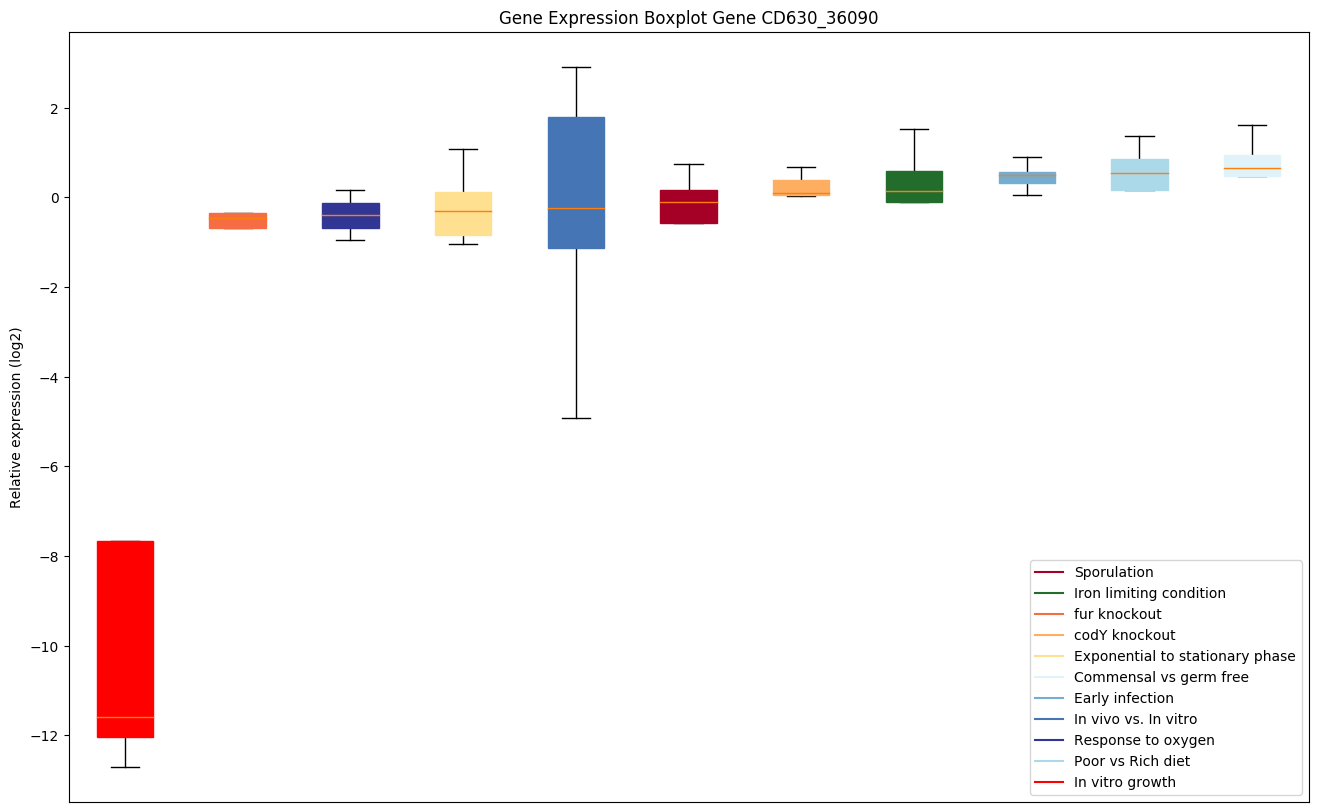 |
|||||
| CD630_27390 | aspS | Aspartyl-tRNA synthetase (Aspartate--tRNA ligase)(AspRS) | Catalyzes the attachment of L-aspartate to tRNA(Asp) in a two-step reaction: L-aspartate is first activated by ATP to form Asp-AMP and then transferred to the acceptor end of tRNA(Asp). | Yes |
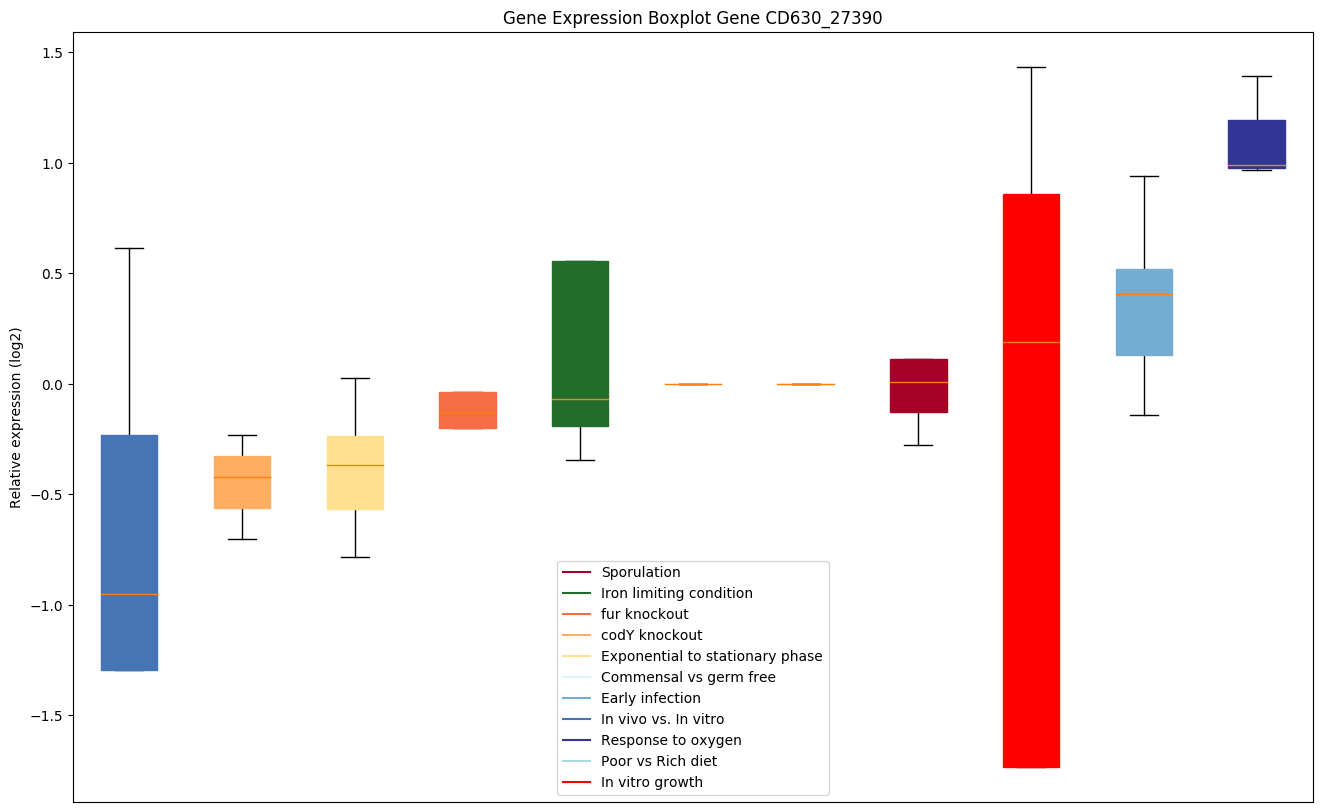 |
||
| CD630_26970 | Putative peptidase, M20D family |
 |
|||||
| CD630_00510 | gltX | Glutamyl-tRNA synthetase | Catalyzes the attachment of glutamate to tRNA(Glu) in a two-step reaction: glutamate is first activated by ATP to form Glu-AMP and then transferred to the acceptor end of tRNA(Glu). | Yes |
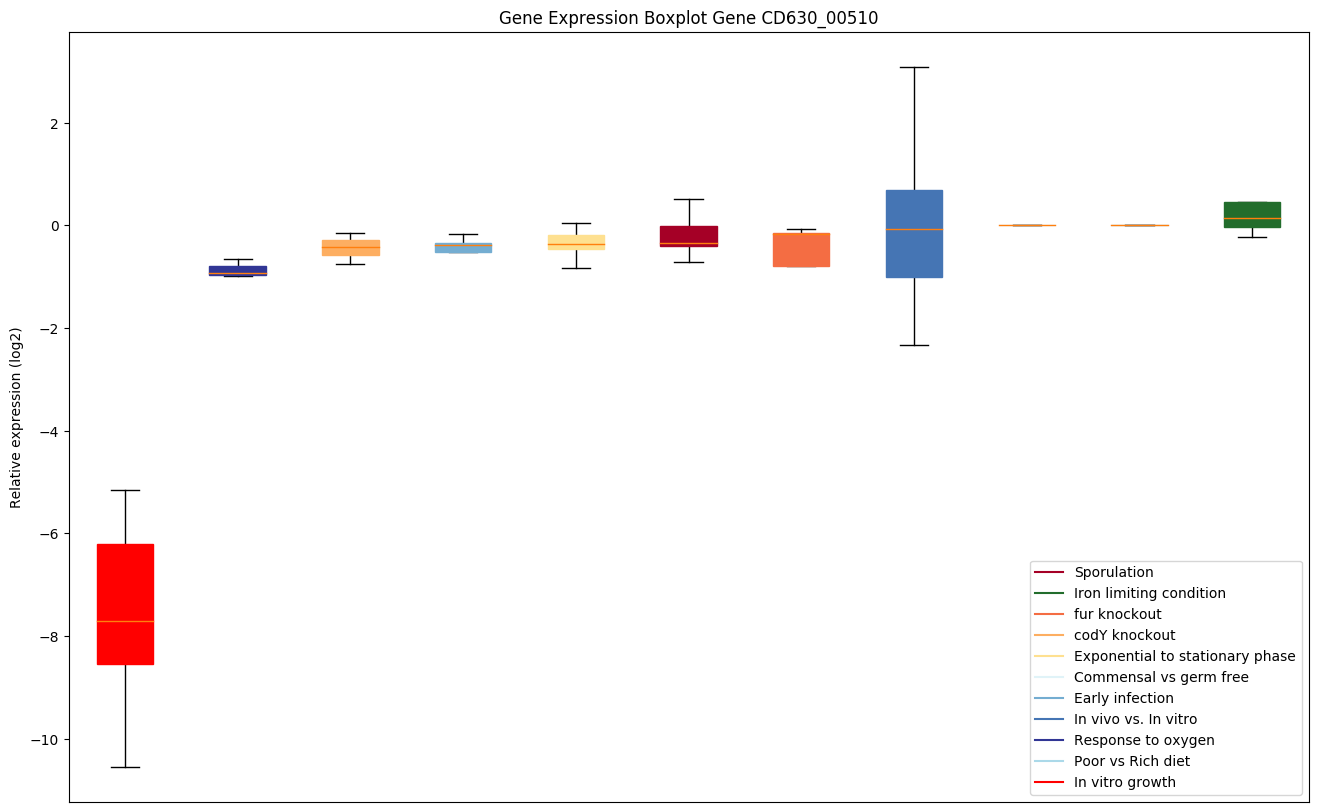 |
||
| CD630_24460 | Putative aspartyl/glutamyl-tRNA amidotransferasesubunit B-related |
 |
|||||
| CD630_27151 | mldC | Midcell localizing division proteins |
 |
||||
| CD630_27170 | mldA | Midcell localizing division proteins |
 |
||||
| CD630_36100 | Conserved hypothetical protein |
 |
|||||
| CD630_33070 | Putative phosphoesterase |
 |
|||||
| CD630_27460 | recJ | Single-stranded-DNA-specific exonuclease |
 |
||||
| CD630_33050 | clpP1 | ATP-dependent Clp protease proteolytic subunit 1 | Cleaves peptides in various proteins in a process that requires ATP hydrolysis. Has a chymotrypsin-like activity. Plays a major role in the degradation of misfolded proteins. |
 |
|||
| CD630_26990 | Putative membrane protein |
 |
|||||
| CD630_05080 | tetM | Tetracycline resistance protein Tn5397 |
 |
||||
| CD630_27160 | mldB | Midcell localizing division proteins |
 |
||||
| CD630_33080 | rph | Bifunctional enzyme, tRNA nucleotidyltransferase; Nucleoside-triphosphatase | Phosphorolytic 3'-5' exoribonuclease that plays an important role in tRNA 3'-end maturation. Removes nucleotide residues following the 3'-CCA terminus of tRNAs; can also add nucleotides to the ends of RNA molecules by using nucleoside diphosphates as substrates, but this may not be physiologically important. Probably plays a role in initiation of 16S rRNA degradation (leading to ribosome degradation) during starvation. |
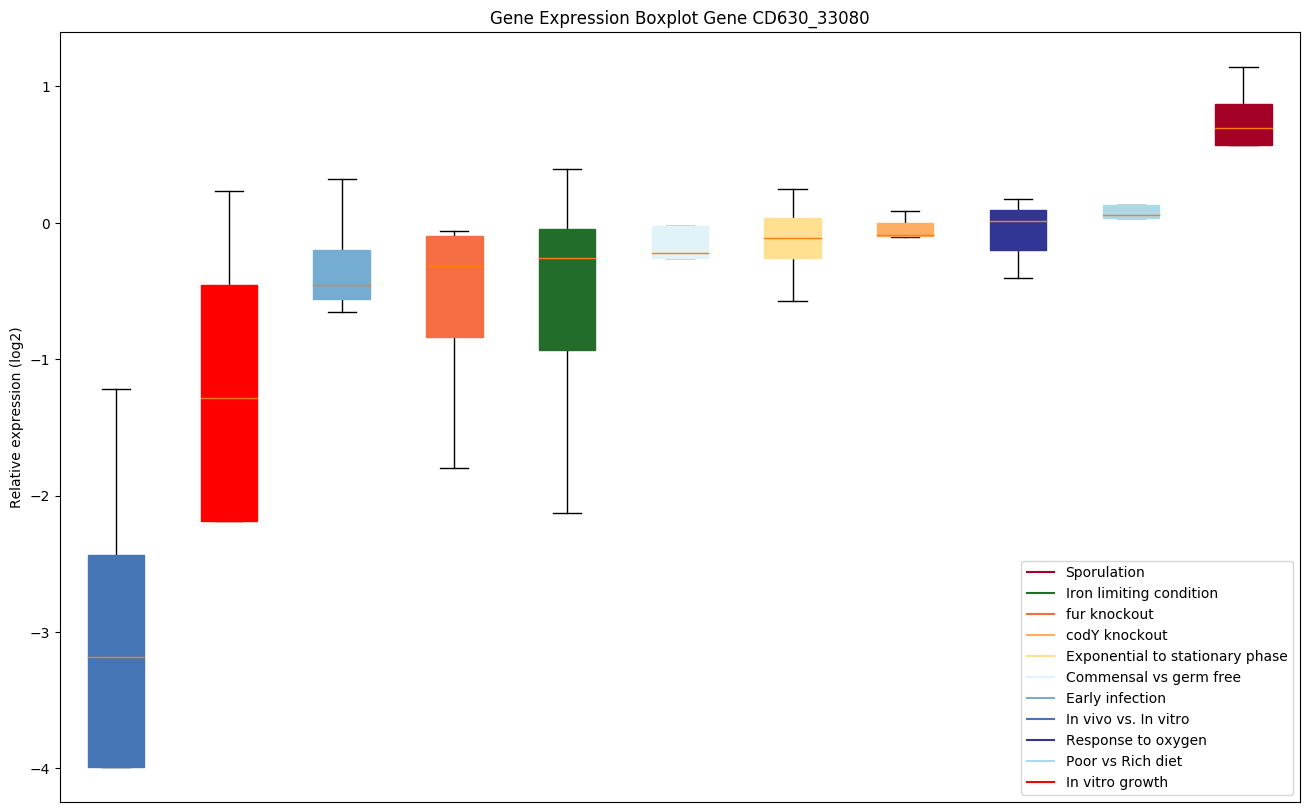 |
|||
| CD630_27410 | hemZ | Coproporphyrinogen III oxidase | Yes |
 |
|||
| CD630_35600 | tilS | tRNA(Ile)-lysidine synthase (tRNA(Ile)-lysidinesynthetase) (tRNA(Ile)-2-lysyl-cytidine synthase) | Ligates lysine onto the cytidine present at position 34 of the AUA codon-specific tRNA(Ile) that contains the anticodon CAU, in an ATP-dependent manner. Cytidine is converted to lysidine, thus changing the amino acid specificity of the tRNA from methionine to isoleucine. | Yes |
 |
||
| CD630_27450 | apt | Adenine phosphoribosyltransferase (APRT) | Catalyzes a salvage reaction resulting in the formation of AMP, that is energically less costly than de novo synthesis. |
 |
|||
| CD630_33060 | tig | Trigger factor (TF) | Involved in protein export. Acts as a chaperone by maintaining the newly synthesized protein in an open conformation. Functions as a peptidyl-prolyl cis-trans isomerase. |
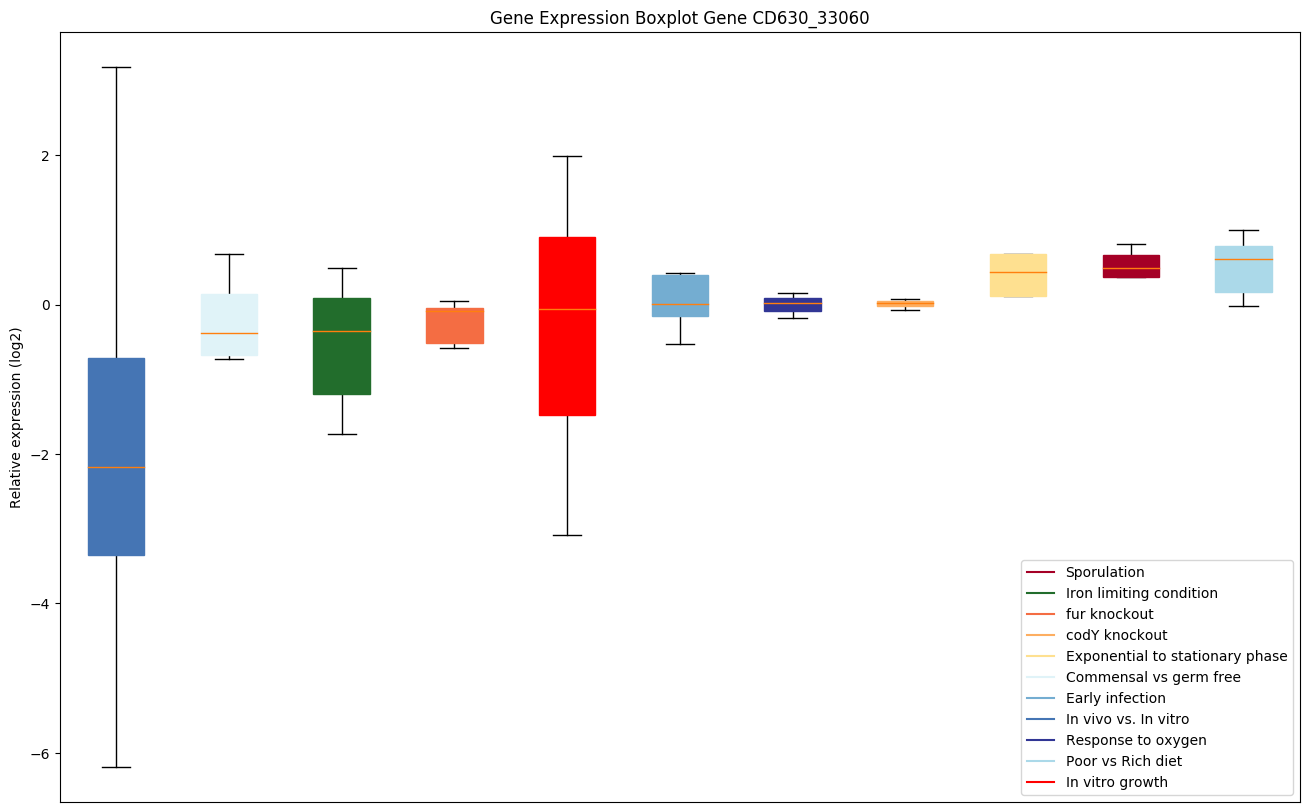 |

