Module 89
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.47 | 27 | 63 | NA | NA |
Bicluster expression profile
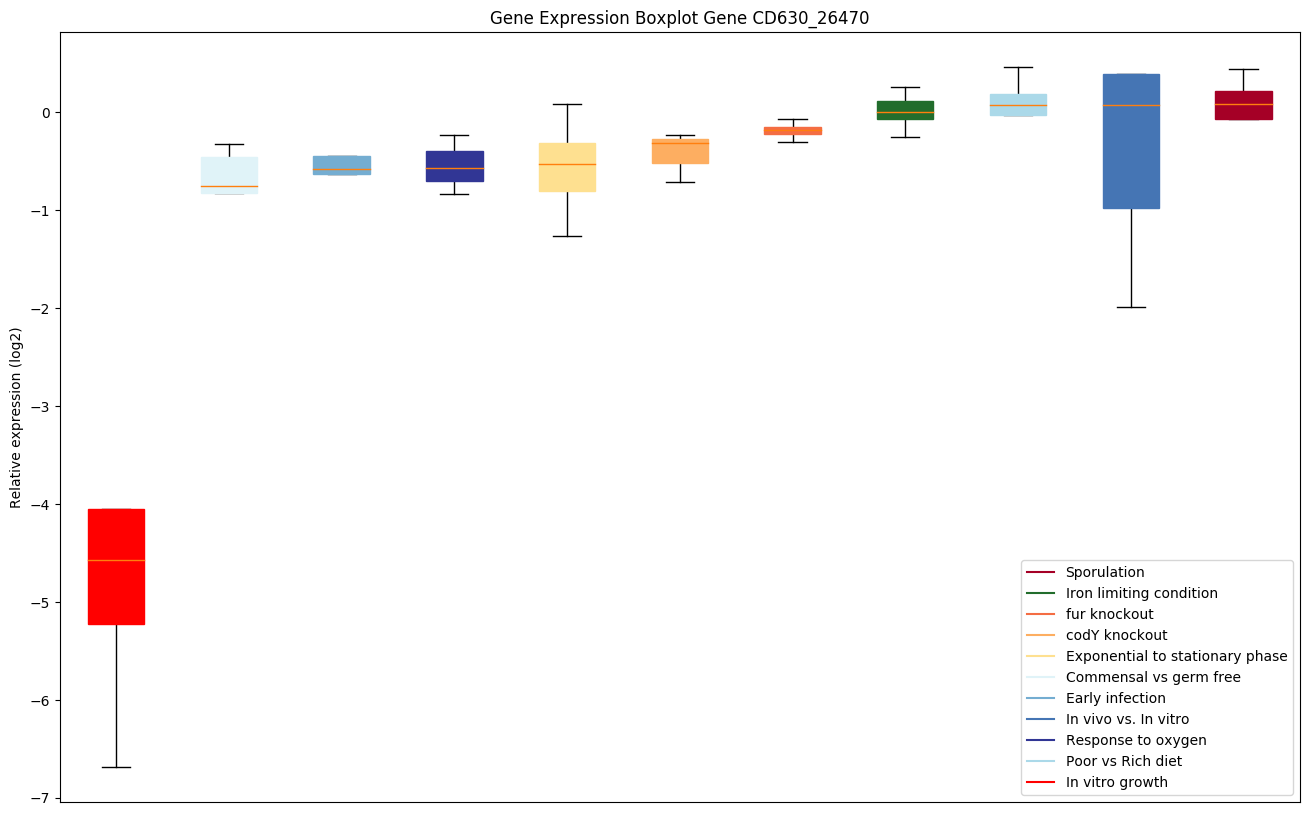
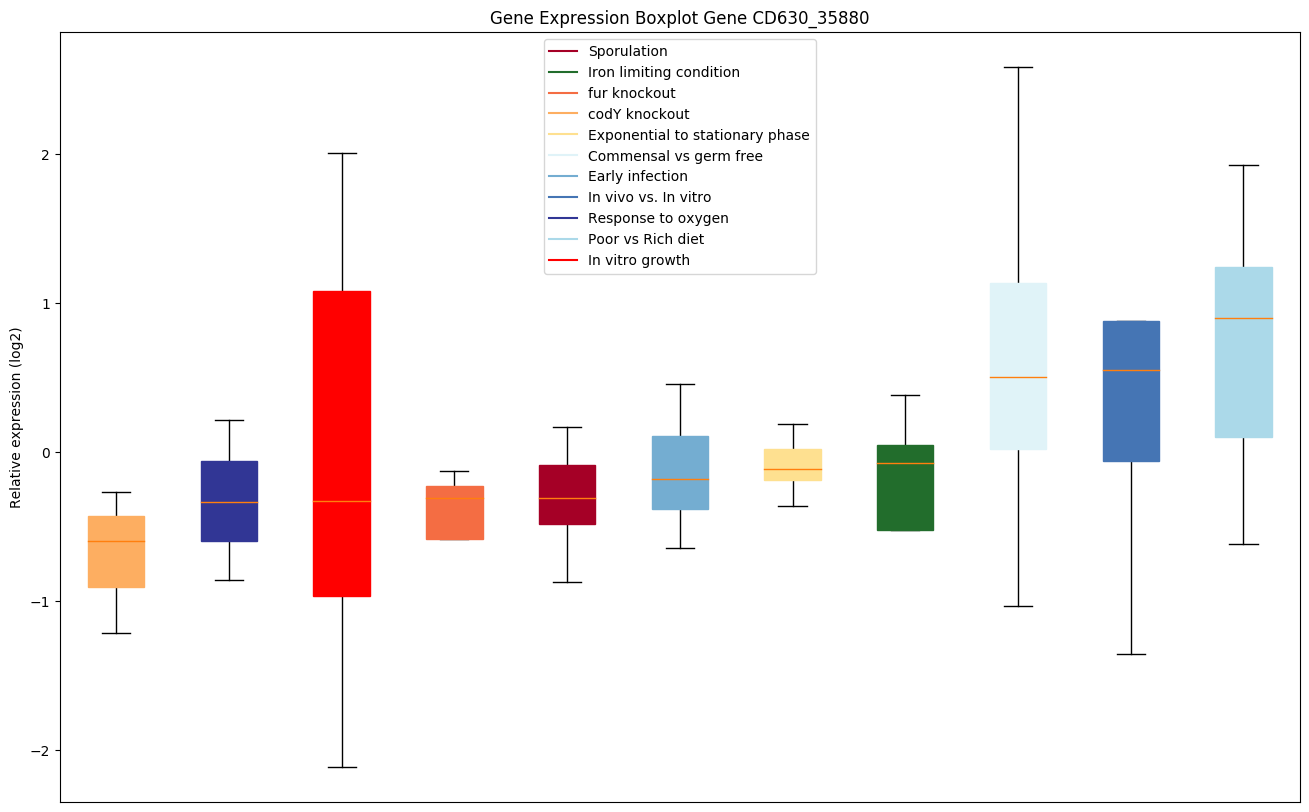
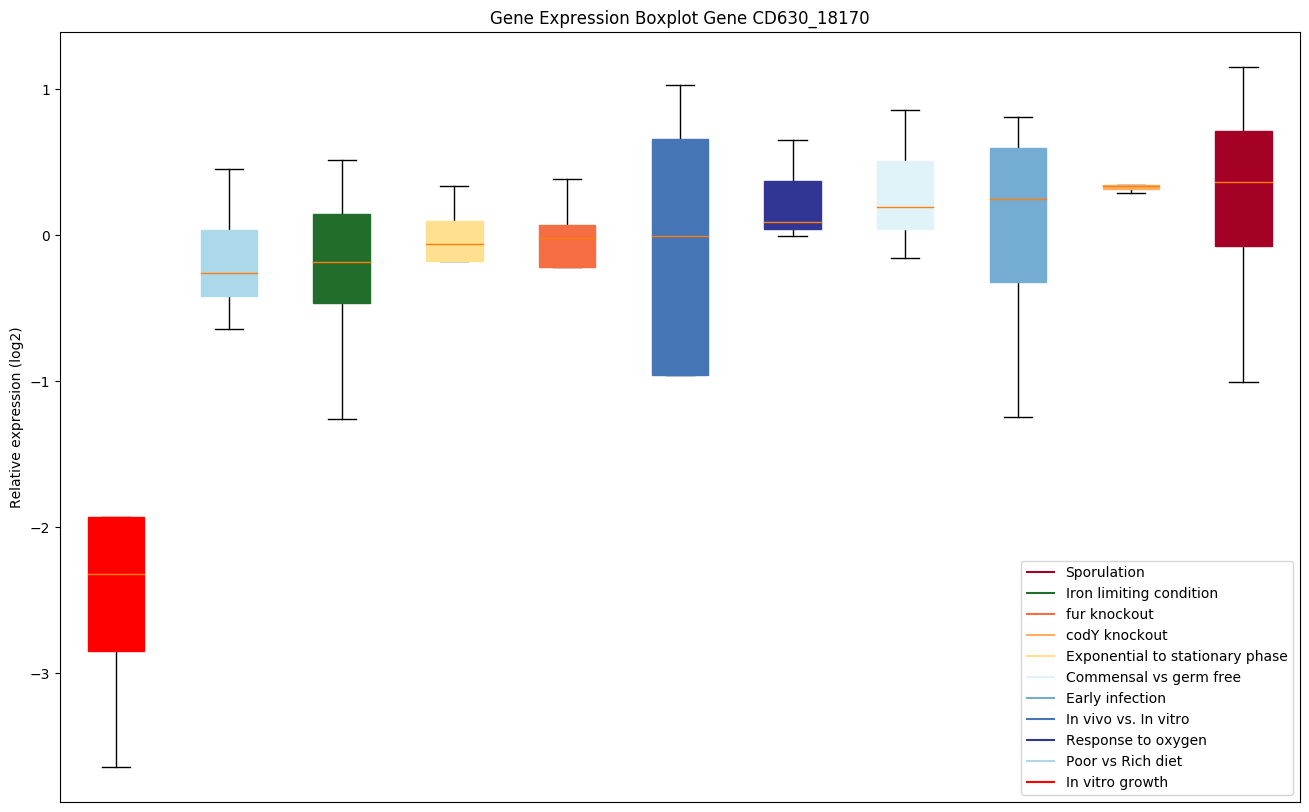
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
| TF | Module | Confidence Score | Beta |
|---|---|---|---|
|
Two-component response regulator |
89 | 0.72 | 0.5554 |
The module is significantly enriched with genes associated to the indicated functional terms
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
| CD630_26550 | murF | UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase (UDP-MurNAc-pentapeptide synthetase)(D-alanyl-D-alanine-adding enzyme) | Involved in cell wall formation. Catalyzes the final step in the synthesis of UDP-N-acetylmuramoyl-pentapeptide, the precursor of murein. | Yes | Yes | Yes |
 |
| CD630_34380 | cobU | Bifunctional adenosylcobalamin biosynthesisprotein CobU [Adenosylcobinamide kinase;Adenosylcobinamide-phosphate guanylyltransferase] |
 |
||||
| CD630_26470 | Putative membrane protein |
 |
|||||
| CD630_18150 | Conserved hypothetical protein |
 |
|||||
| CD630_26490 | Conserved hypothetical protein |
 |
|||||
| CD630_35880 | carB1 | Carbamoyl-phosphate synthase large chain 1 |
 |
||||
| CD630_18170 | Putative acyltransferase | Yes |
 |
||||
| CD630_35400 | metG | Methionyl-tRNA synthetase (Methionine--tRNAligase) (MetRS) | Is required not only for elongation of protein synthesis but also for the initiation of all mRNA translation through initiator tRNA(fMet) aminoacylation. | Yes |
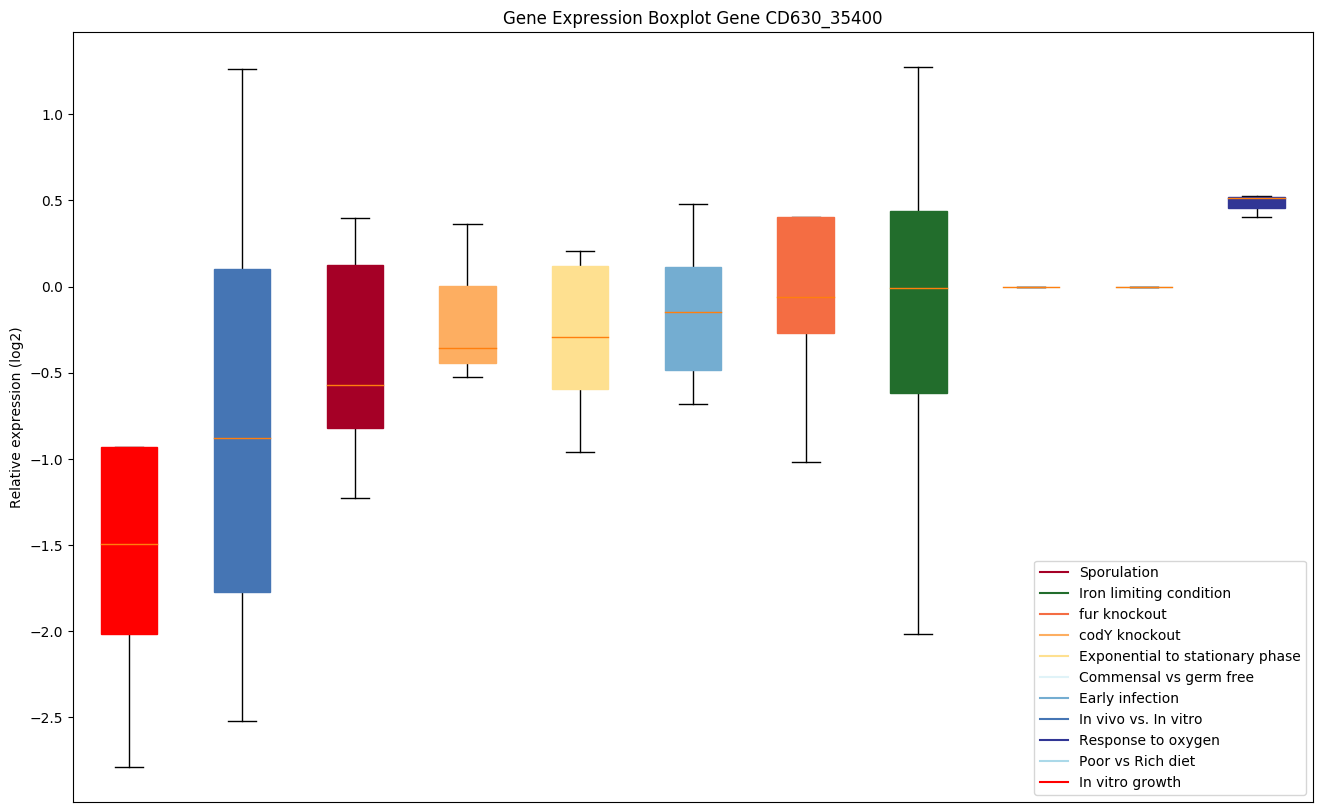 |
||
| CD630_34370 | cobS | Cobalamin synthase CobS (Cobalamin-5-phosphatesynthase) | Joins adenosylcobinamide-GDP and alpha-ribazole to generate adenosylcobalamin (Ado-cobalamin). Also synthesizes adenosylcobalamin 5'-phosphate from adenosylcobinamide-GDP and alpha-ribazole 5'-phosphate. |
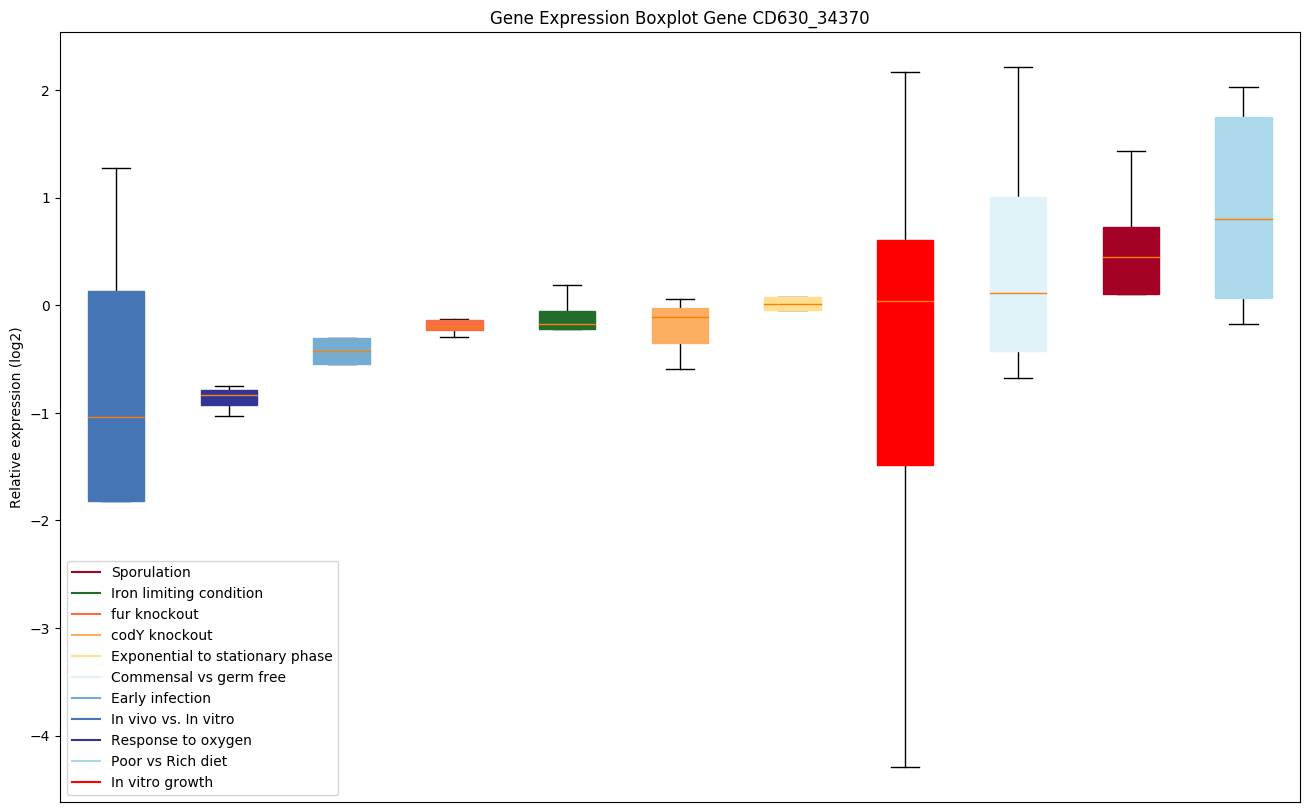 |
|||
| CD630_26520 | spoVE | Cell division/stage V sporulation protein |
 |
||||
| CD630_35390 | Putative deoxyribonuclease |
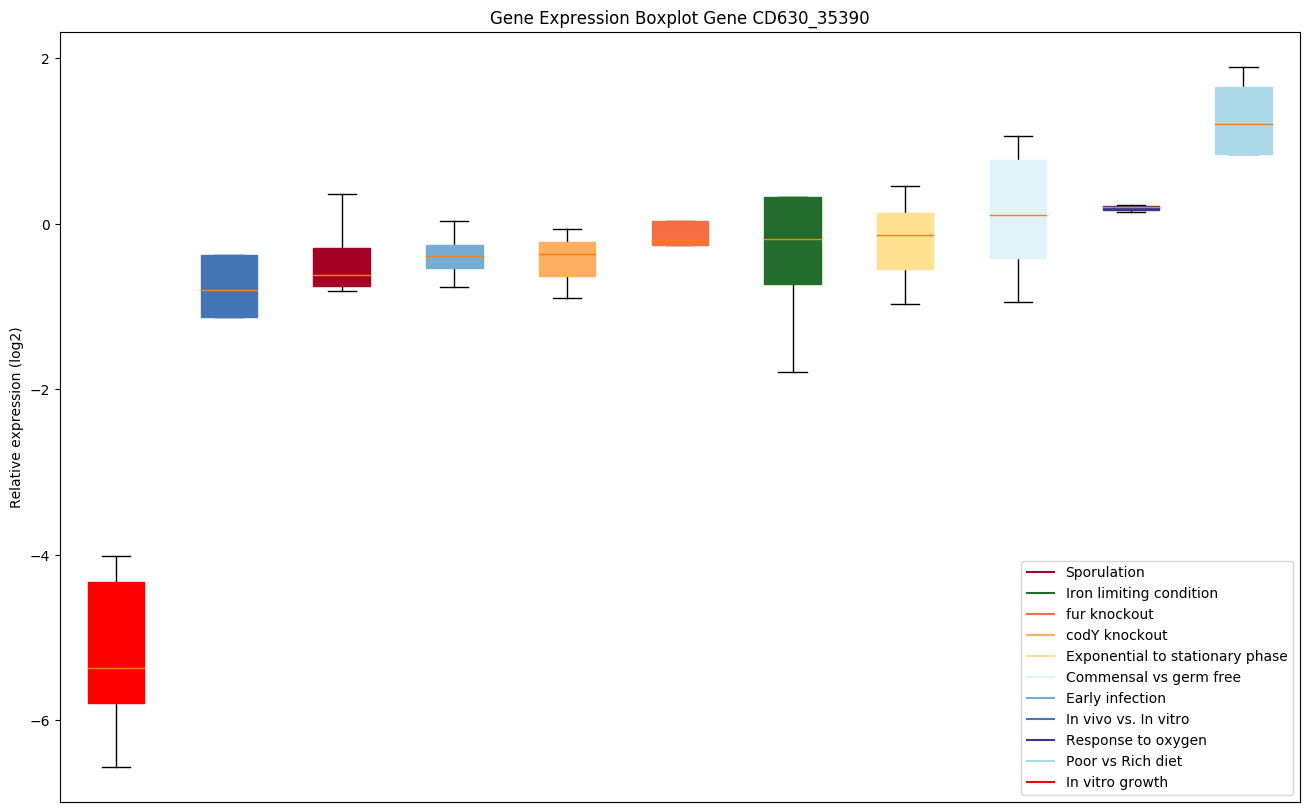 |
|||||
| CD630_18140 | Putative phosphoglycerate mutase |
 |
|||||
| CD630_35910 | carA | Carbamoyl-phosphate synthase small chain |
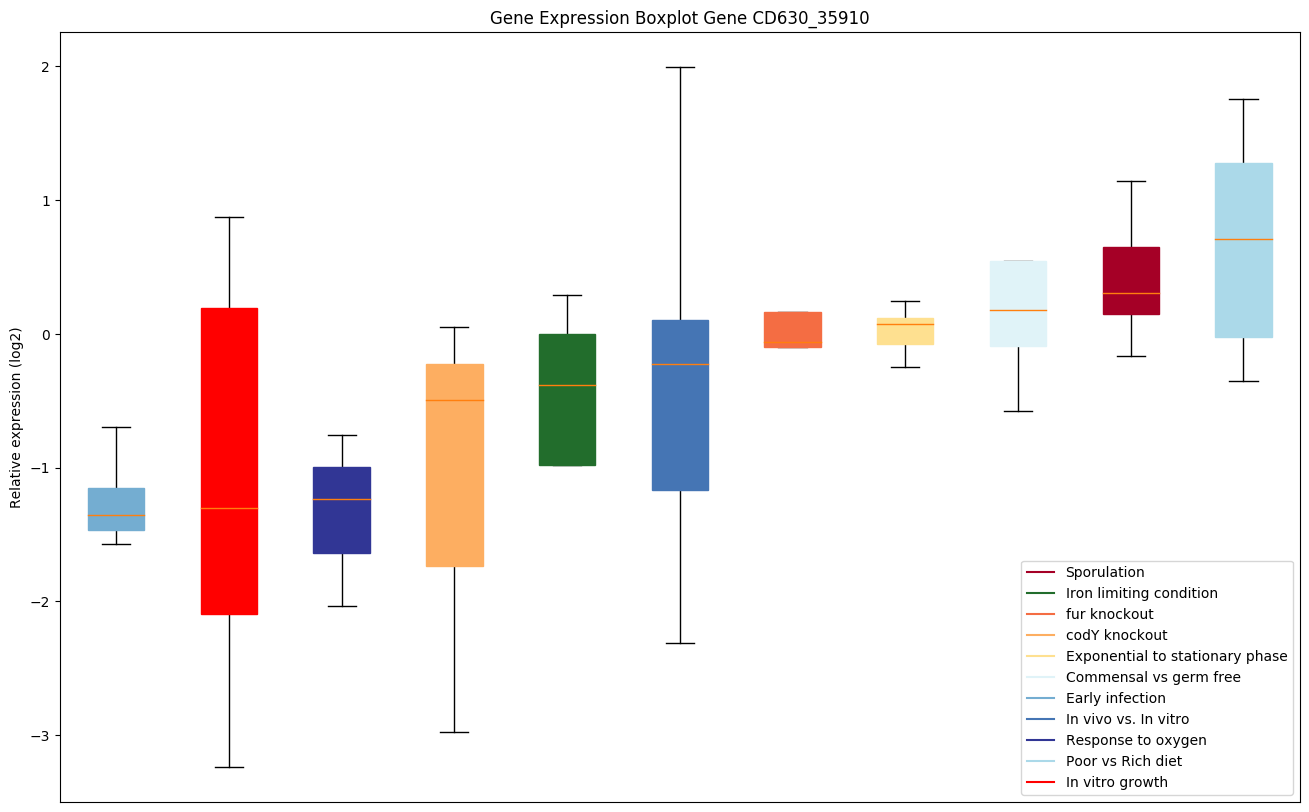 |
||||
| CD630_36250 | Putative signaling protein |
 |
|||||
| CD630_26500 | Putative cell division protein Fts-Q type | Essential cell division protein. |
 |
||||
| CD630_18160 | cmk | Cytidylate kinase (CK) (Cytidine monophosphatekinase) (CMP kinase) | Yes |
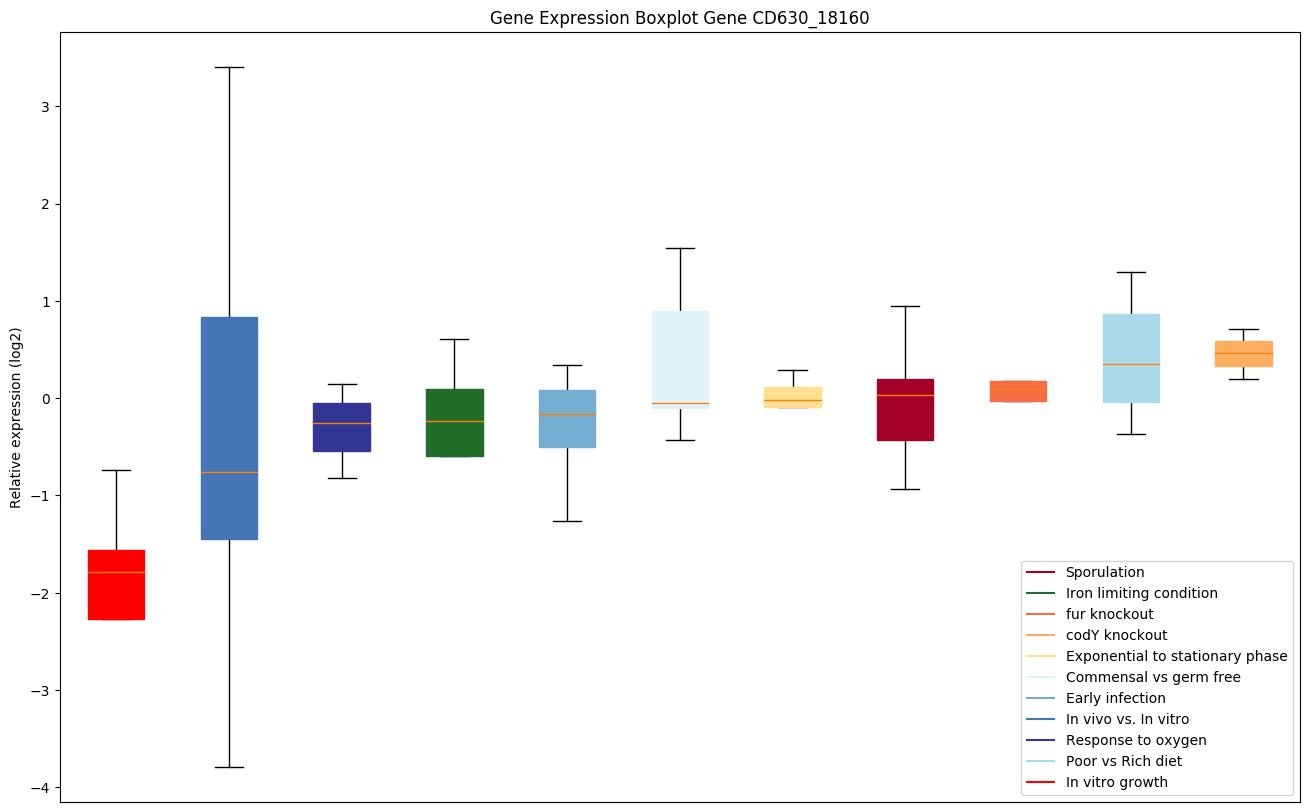 |
|||
| CD630_26540 | mraY | Phospho-N-acetylmuramoyl-pentapeptide-transferase(UDP- MurNAc-pentapeptide phosphotransferase) | First step of the lipid cycle reactions in the biosynthesis of the cell wall peptidoglycan. | Yes |
 |
||
| CD630_35890 | carA | Carbamoyl-phosphate synthase small chain |
 |
||||
| CD630_34390 | cobT | Nicotinate-nucleotide--dimethylbenzimidazolephosphoribosyltransferase (NN:DBI PRT)(N(1)-alpha-phosphoribosyltransferase) | Catalyzes the synthesis of alpha-ribazole-5'-phosphate from nicotinate mononucleotide (NAMN) and 5,6-dimethylbenzimidazole (DMB). |
 |
|||
| CD630_26480 | Conserved hypothetical protein |
 |
|||||
| CD630_24800 | Putative hydrolase |
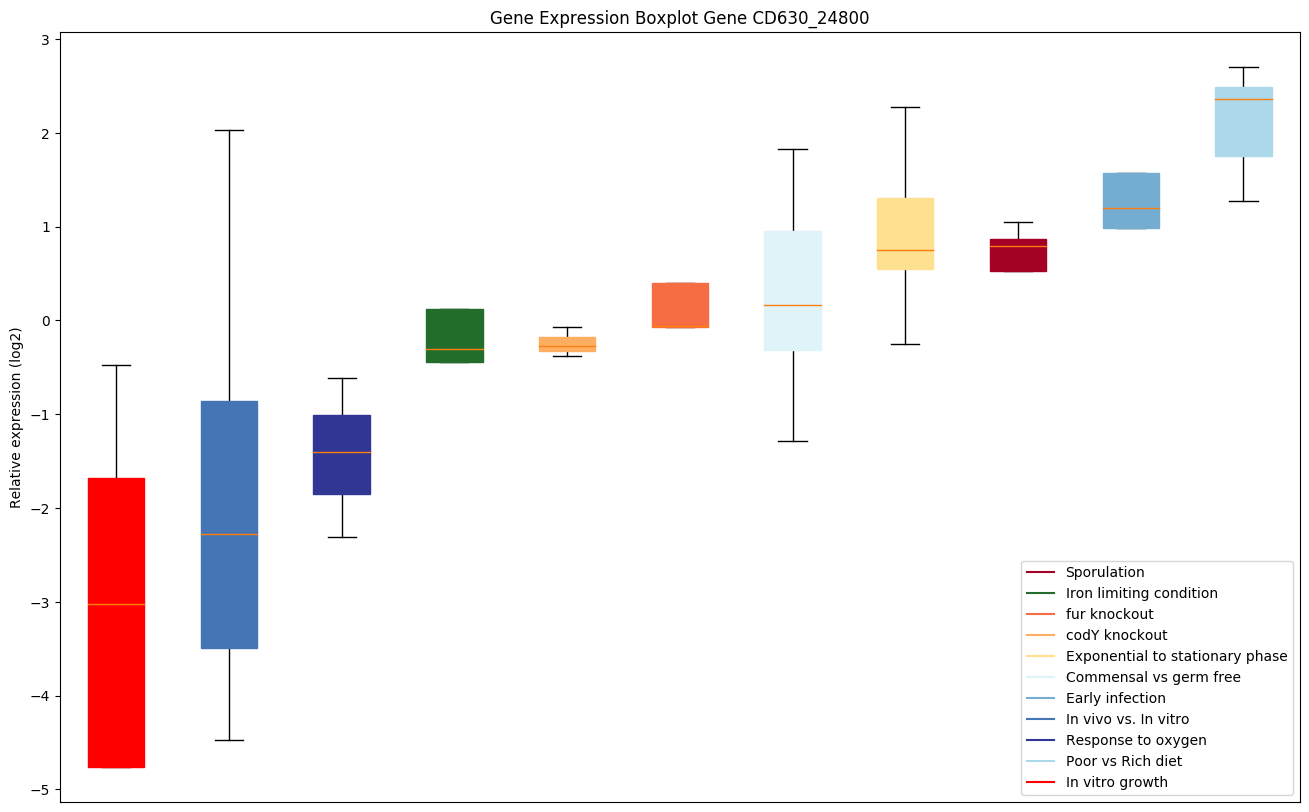 |
|||||
| CD630_18180 | ispH | 4-hydroxy-3-methylbut-2-enyl diphosphatereductase | Catalyzes the conversion of 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (HMBPP) into a mixture of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). Acts in the terminal step of the DOXP/MEP pathway for isoprenoid precursor biosynthesis. | Yes | Yes | Yes |
 |
| CD630_34360 | cobC | Cobalamin biosynthesis phosphoglycerate mutaseCobC |
 |
||||
| CD630_35920 | pyrF | Orotidine 5'-phosphate decarboxylase (OMPdecarboxylase) (OMPDCase) (OMPdecase) | Catalyzes the decarboxylation of orotidine 5'-monophosphate (OMP) to uridine 5'-monophosphate (UMP). |
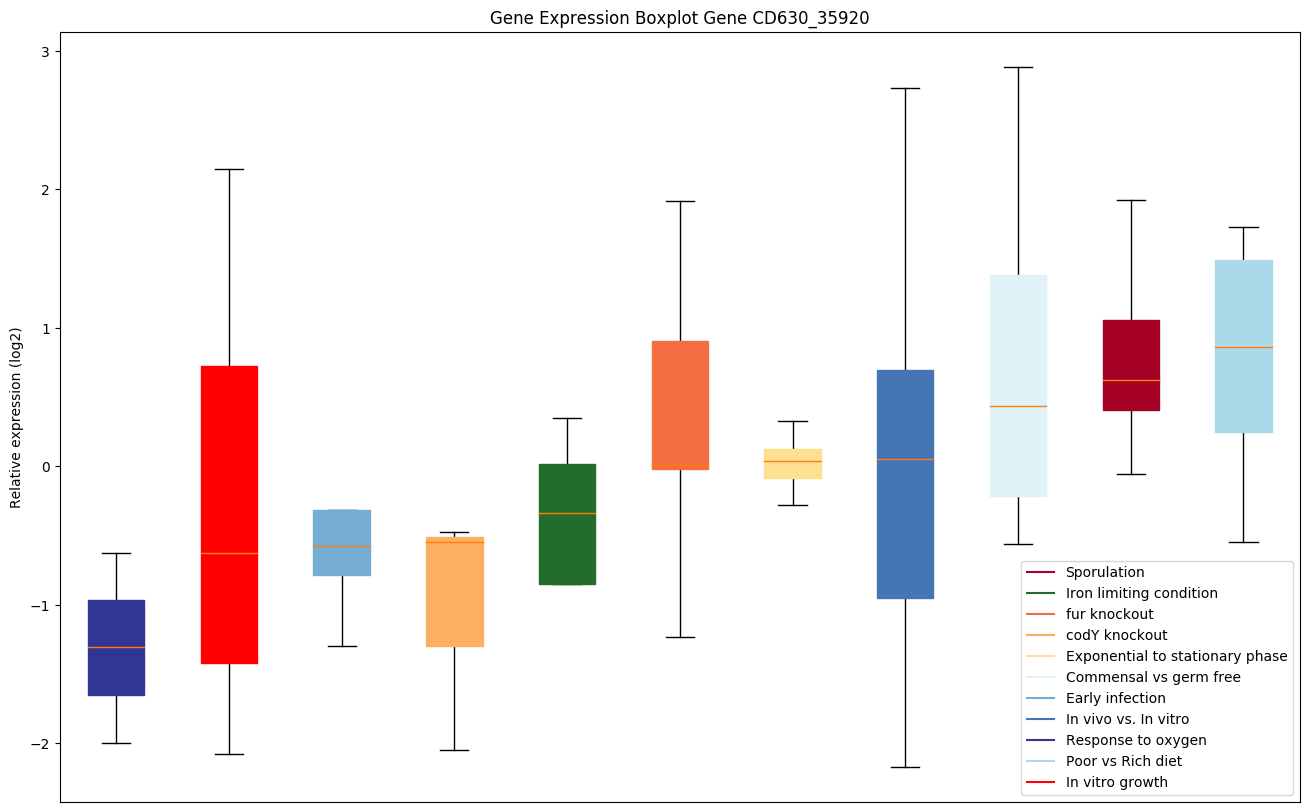 |
|||
| CD630_26530 | murD | UDP-N-acetylmuramoylalanine--D-glutamate ligase(UDP-N- acetylmuramoyl-L-alanyl-D-glutamate synthetase)(D-glutamic acid- adding enzyme) | Cell wall formation. Catalyzes the addition of glutamate to the nucleotide precursor UDP-N-acetylmuramoyl-L-alanine (UMA). | Yes | Yes | Yes |
 |
| CD630_26510 | murG | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosaminetransferase | Cell wall formation. Catalyzes the transfer of a GlcNAc subunit on undecaprenyl-pyrophosphoryl-MurNAc-pentapeptide (lipid intermediate I) to form undecaprenyl-pyrophosphoryl-MurNAc-(pentapeptide)GlcNAc (lipid intermediate II). | Yes | Yes | Yes |
 |
| CD630_35900 | carB2 | Carbamoyl-phosphate synthase large chain 2 |
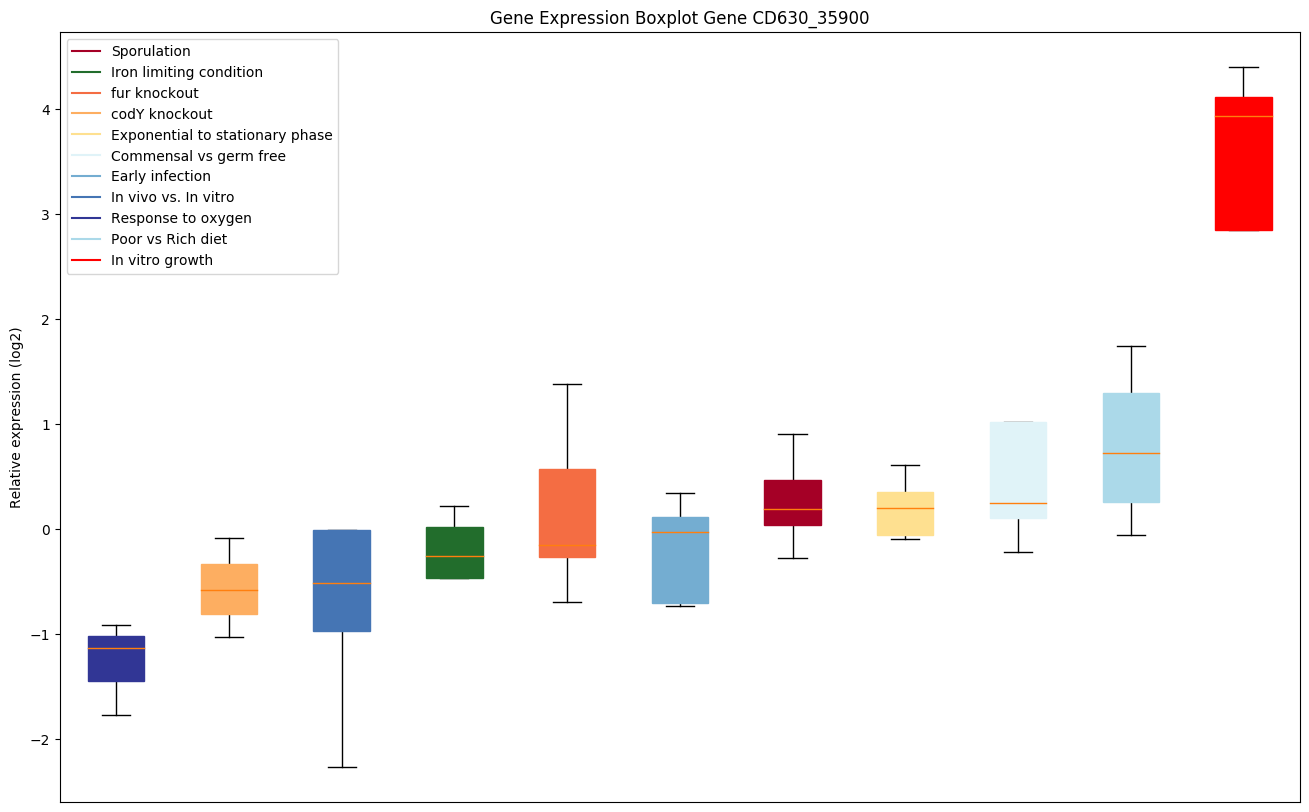 |