Module 192
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.47 | 23 | 65 | NA | NA |
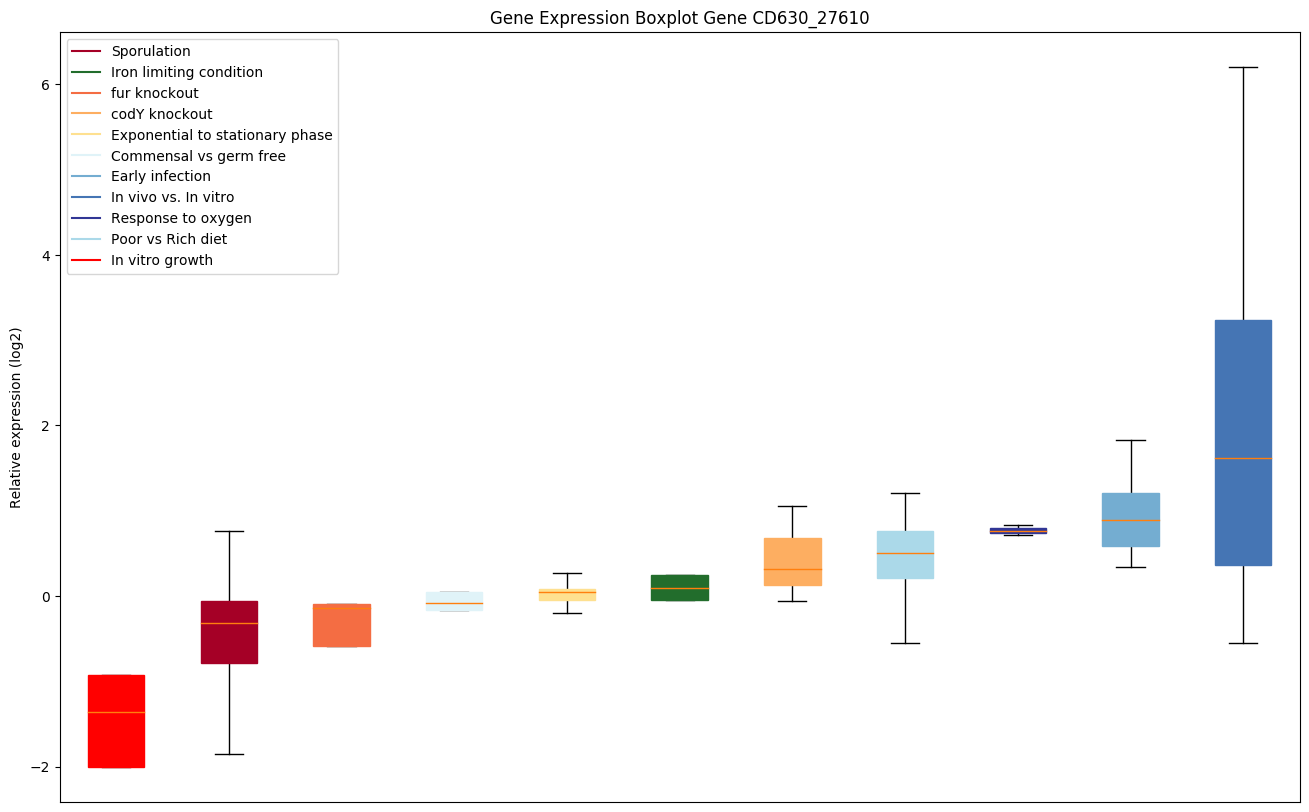
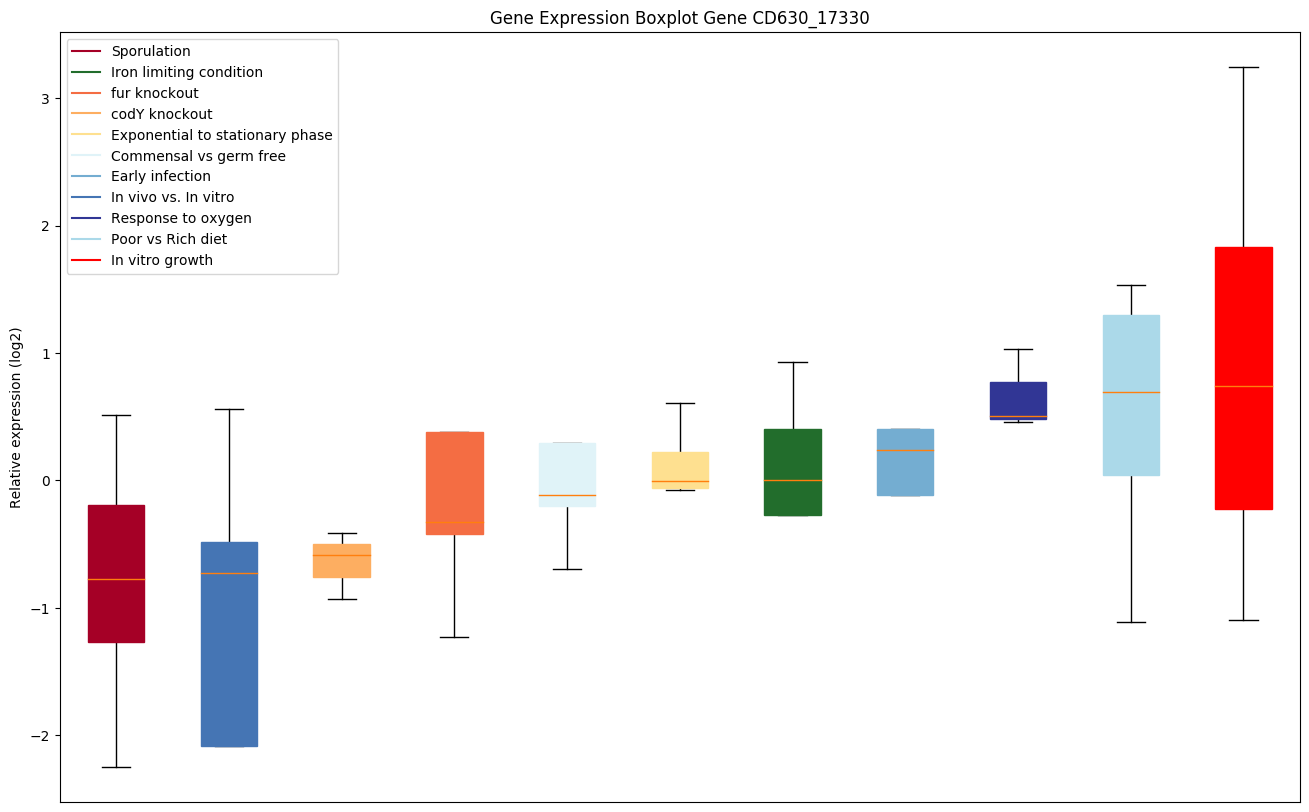
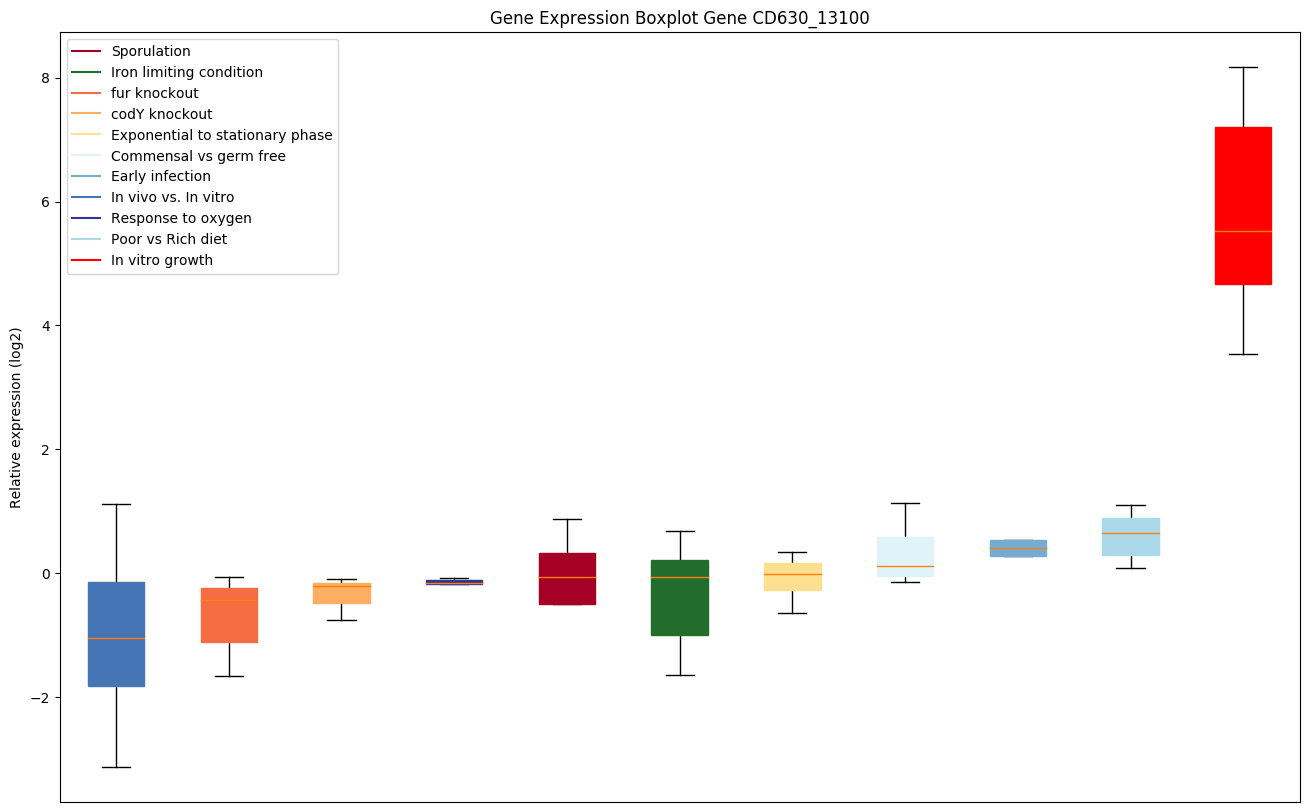
Bicluster expression profile
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
| TF | Module | Confidence Score | Beta |
|---|---|---|---|
|
Two-component response regulator |
192 | 0.18 | -0.1039 |
|
Transcriptional regulator, beta-lactamsrepressor |
192 | 0.15 | 0.1128 |
|
Transcriptional regulator, DeoR family |
192 | 0.18 | 0.1090 |
|
Transcriptional regulator, beta-lactamsrepressor |
192 | 0.25 | 0.1258 |
|
Transcriptional regulator, GntR family |
192 | 0.23 | 0.1475 |
The module is significantly enriched with genes associated to the indicated functional terms
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
| CD630_27610 | Putative N-acetylmuramoyl-L-alanine amidase |
 |
|||||
| CD630_36550 | purA | Adenylosuccinate synthetase (IMP--aspartateligase) (AdSS) (AMPSase) | Plays an important role in the de novo pathway of purine nucleotide biosynthesis. Catalyzes the first committed step in the biosynthesis of AMP from IMP. |
 |
|||
| CD630_17330 | hemE | Uroporphyrinogen decarboxylase (URO-D) |
 |
||||
| CD630_13100 | rbfA | Ribosome-binding factor A | One of several proteins that assist in the late maturation steps of the functional core of the 30S ribosomal subunit. Associates with free 30S ribosomal subunits (but not with 30S subunits that are part of 70S ribosomes or polysomes). Required for efficient processing of 16S rRNA. May interact with the 5'-terminal helix region of 16S rRNA. |
 |
|||
| CD630_13060 | Conserved hypothetical protein | Required for maturation of 30S ribosomal subunits. |
 |
||||
| CD630_13080 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 | Yes |
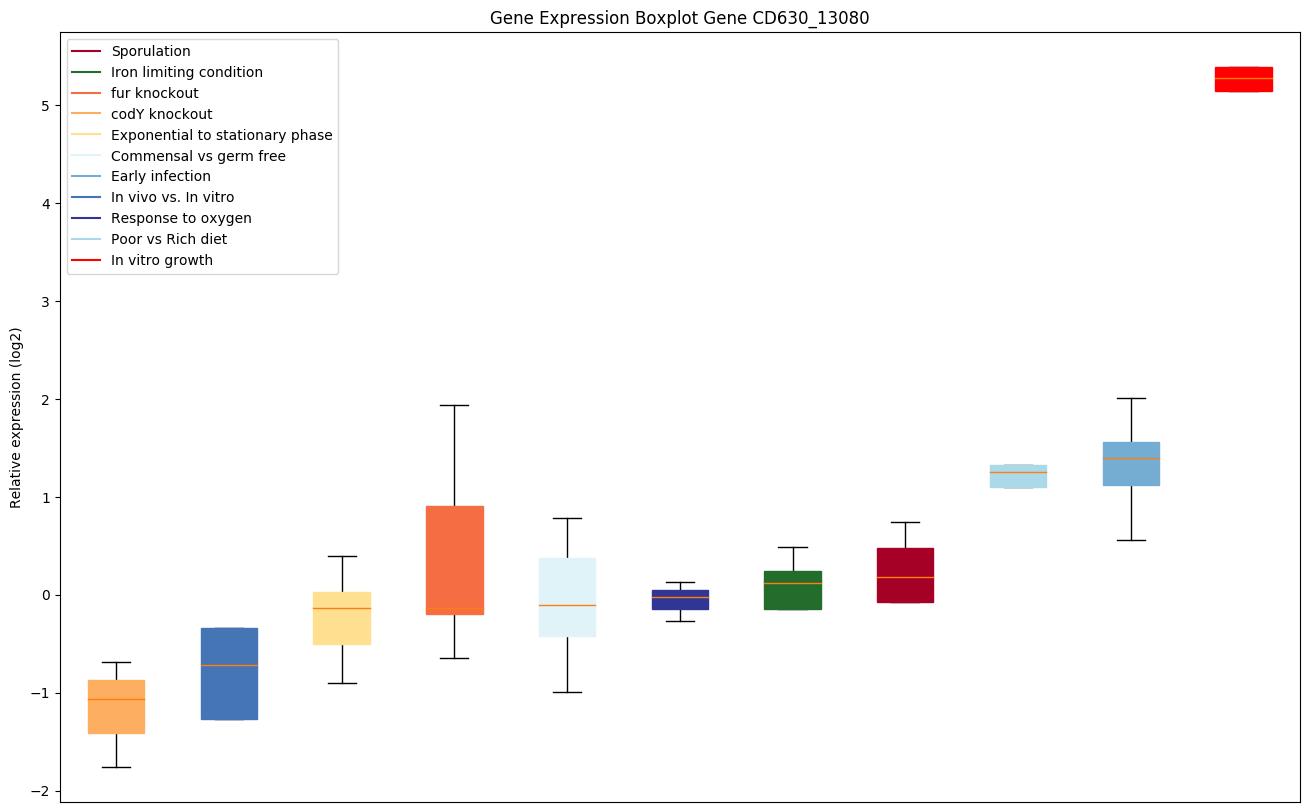 |
||||
| CD630_22750 | topB2 | DNA topoisomerase III | Releases the supercoiling and torsional tension of DNA, which is introduced during the DNA replication and transcription, by transiently cleaving and rejoining one strand of the DNA duplex. Introduces a single-strand break via transesterification at a target site in duplex DNA. The scissile phosphodiester is attacked by the catalytic tyrosine of the enzyme, resulting in the formation of a DNA-(5'-phosphotyrosyl)-enzyme intermediate and the expulsion of a 3'-OH DNA strand. The free DNA strand then undergoes passage around the unbroken strand, thus removing DNA supercoils. Finally, in the religation step, the DNA 3'-OH attacks the covalent intermediate to expel the active-site tyrosine and restore the DNA phosphodiester backbone. |
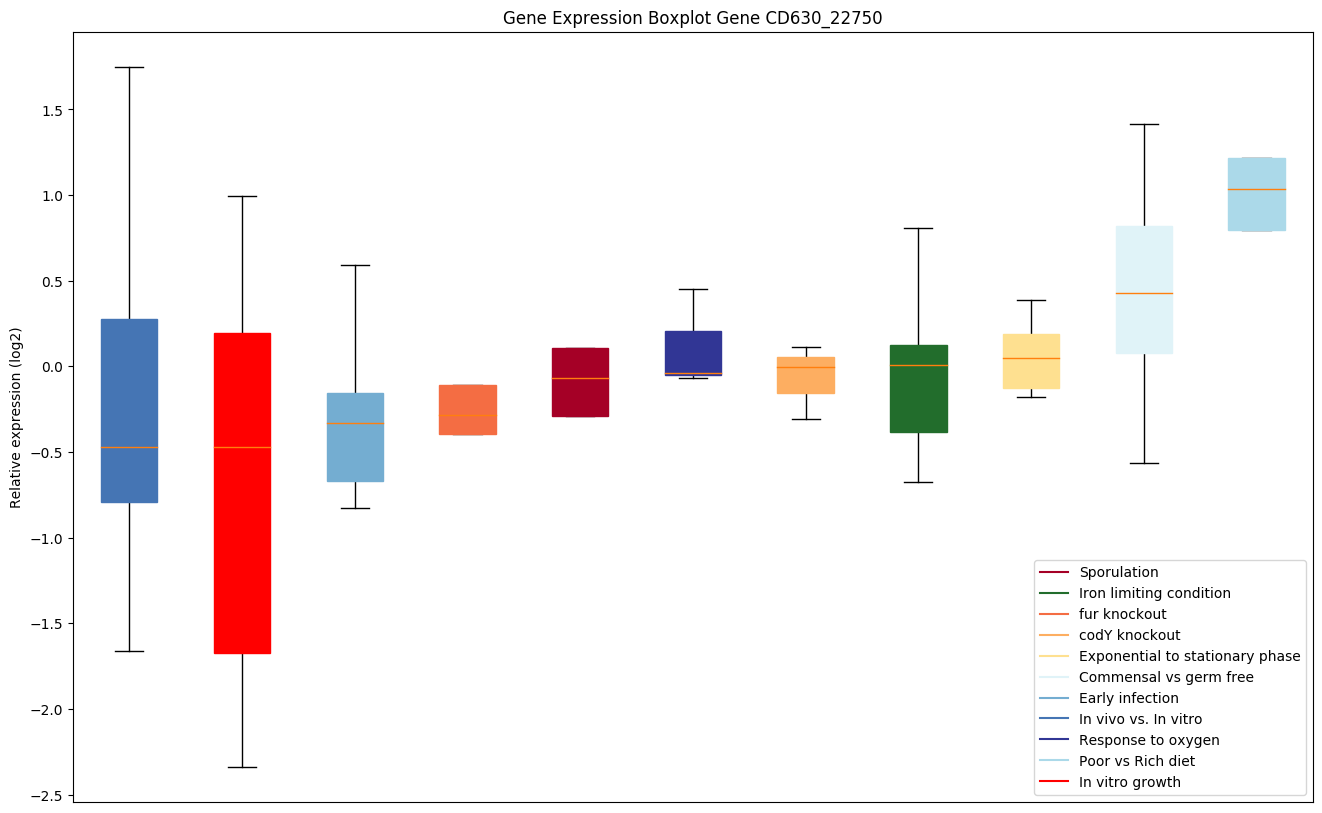 |
|||
| CD630_07100 | Conserved hypothetical protein, DUF523 family |
 |
|||||
| CD630_28020 | tgt | Queuine tRNA-ribosyltransferase (tRNA-guaninetransglycosylase) (Guanine insertion enzyme) | Catalyzes the base-exchange of a guanine (G) residue with the queuine precursor 7-aminomethyl-7-deazaguanine (PreQ1) at position 34 (anticodon wobble position) in tRNAs with GU(N) anticodons (tRNA-Asp, -Asn, -His and -Tyr). Catalysis occurs through a double-displacement mechanism. The nucleophile active site attacks the C1' of nucleotide 34 to detach the guanine base from the RNA, forming a covalent enzyme-RNA intermediate. The proton acceptor active site deprotonates the incoming PreQ1, allowing a nucleophilic attack on the C1' of the ribose to form the product. After dissociation, two additional enzymatic reactions on the tRNA convert PreQ1 to queuine (Q), resulting in the hypermodified nucleoside queuosine (7-(((4,5-cis-dihydroxy-2-cyclopenten-1-yl)amino)methyl)-7-deazaguanosine). |
 |
|||
| CD630_28000 | Putative membrane protein |
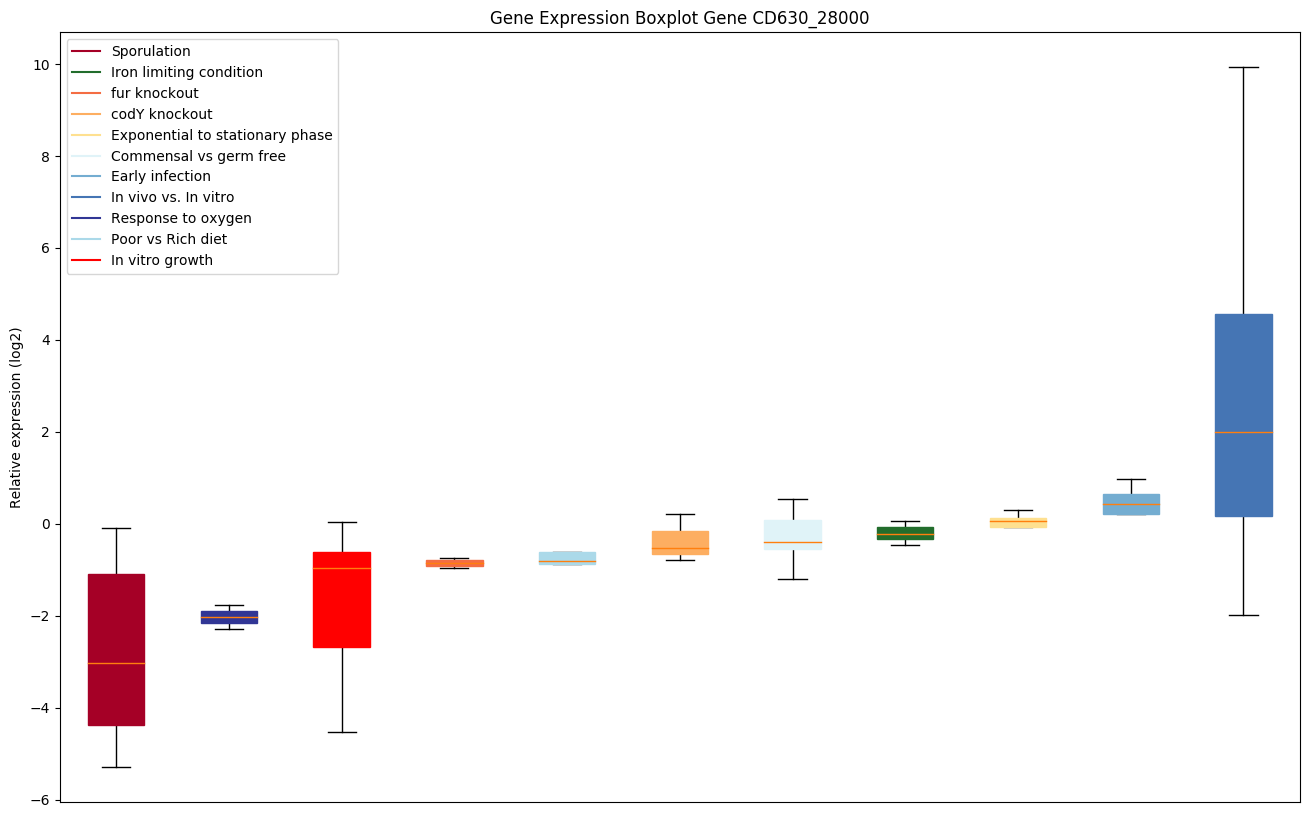 |
|||||
| CD630_28040 | queA | S-adenosylmethionine:tRNAribosyltransferase-isomerase (Queuosine biosynthesisprotein QueA) | Transfers and isomerizes the ribose moiety from AdoMet to the 7-aminomethyl group of 7-deazaguanine (preQ1-tRNA) to give epoxyqueuosine (oQ-tRNA). |
 |
|||
| CD630_17320 | Putative cobalamin-binding protein |
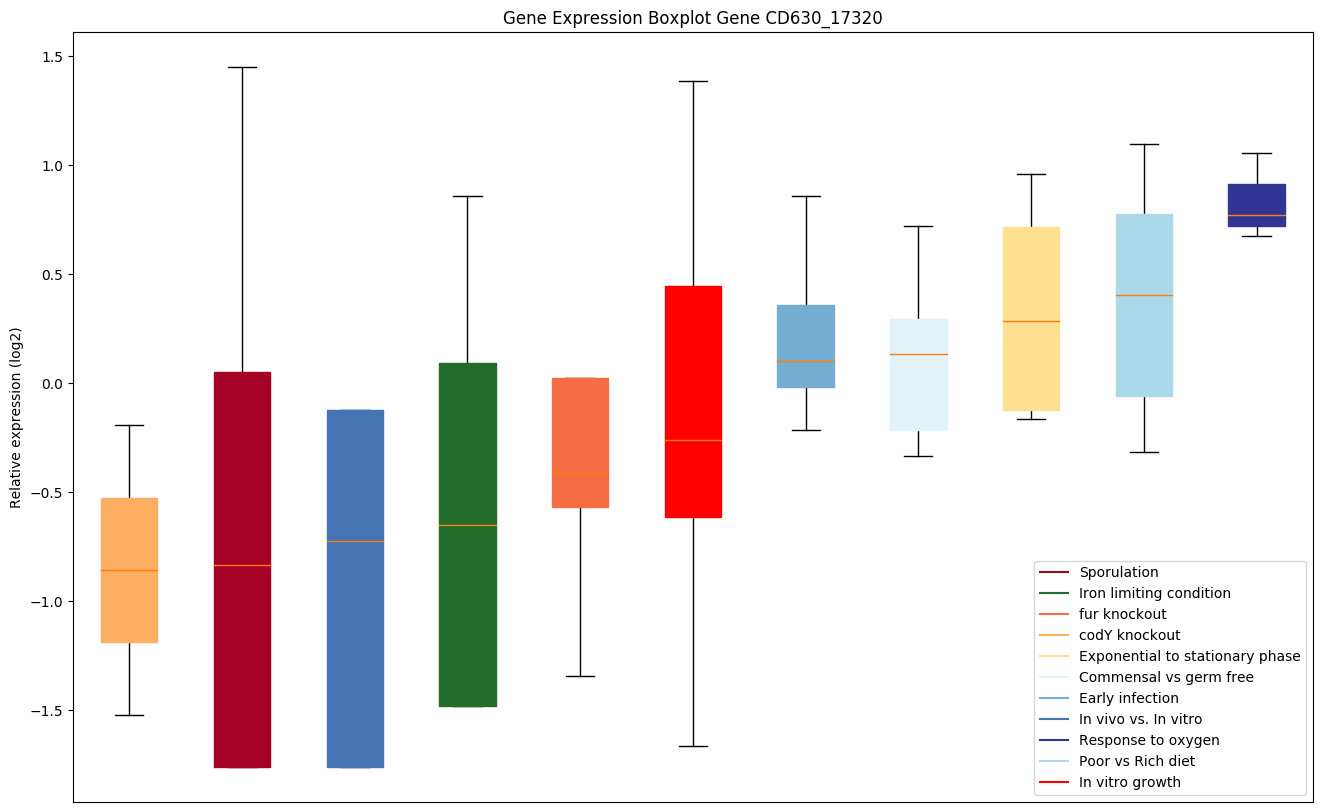 |
|||||
| CD630_13071 | Conserved hypothetical protein | Yes |
 |
||||
| CD630_13070 | nusA | Transcription elongation protein | Participates in both transcription termination and antitermination. | Yes |
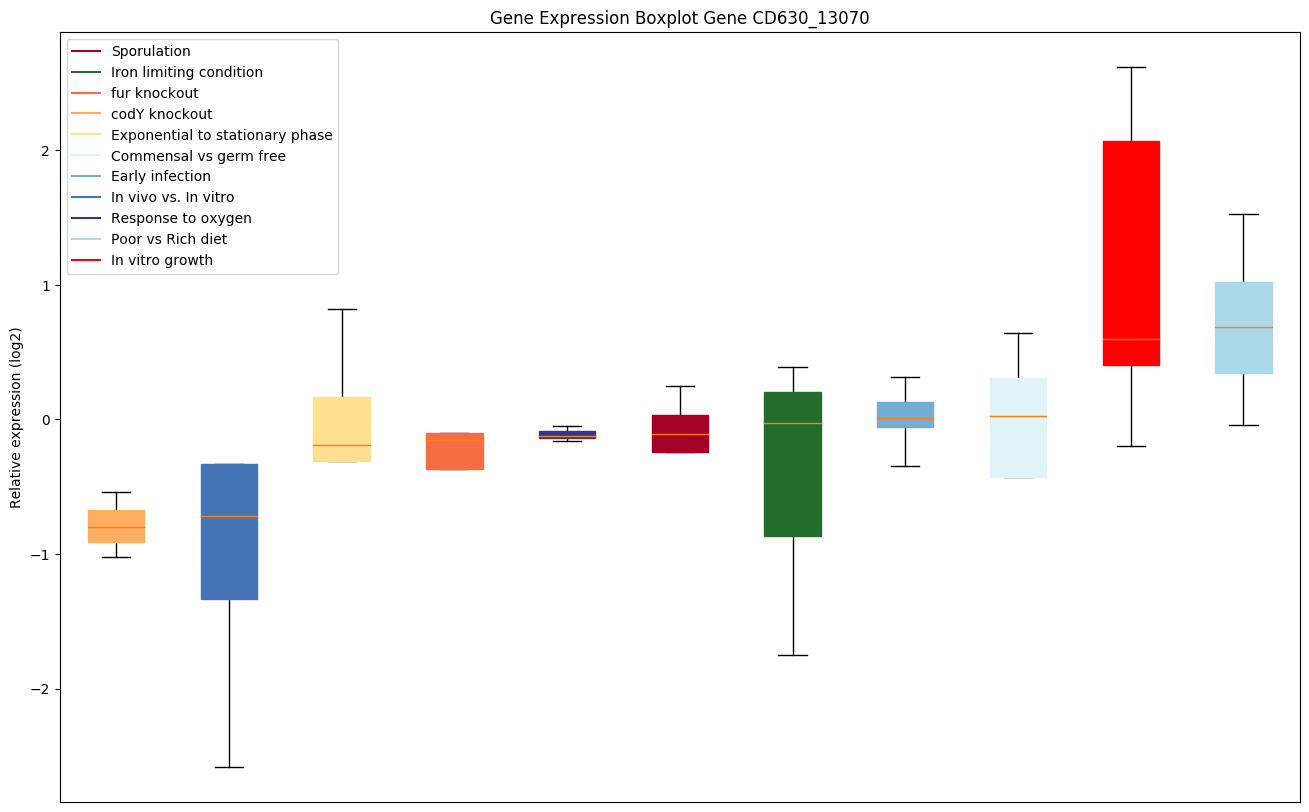 |
||
| CD630_36560 | Conserved hypothetical protein with a TonB box |
 |
|||||
| CD630_17340 | Conserved hypothetical protein |
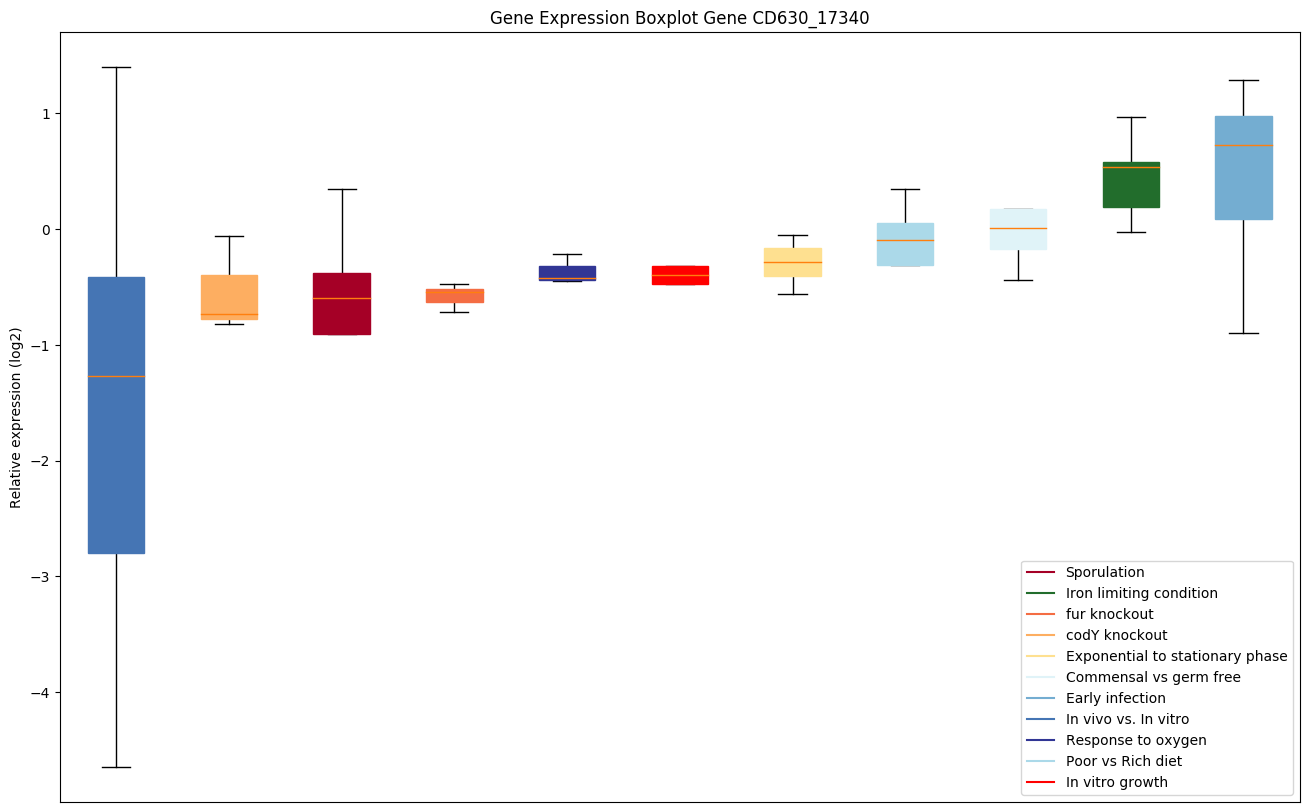 |
|||||
| CD630_13090 | infB | Translation initiation factor IF-2 | One of the essential components for the initiation of protein synthesis. Protects formylmethionyl-tRNA from spontaneous hydrolysis and promotes its binding to the 30S ribosomal subunits. Also involved in the hydrolysis of GTP during the formation of the 70S ribosomal complex. | Yes |
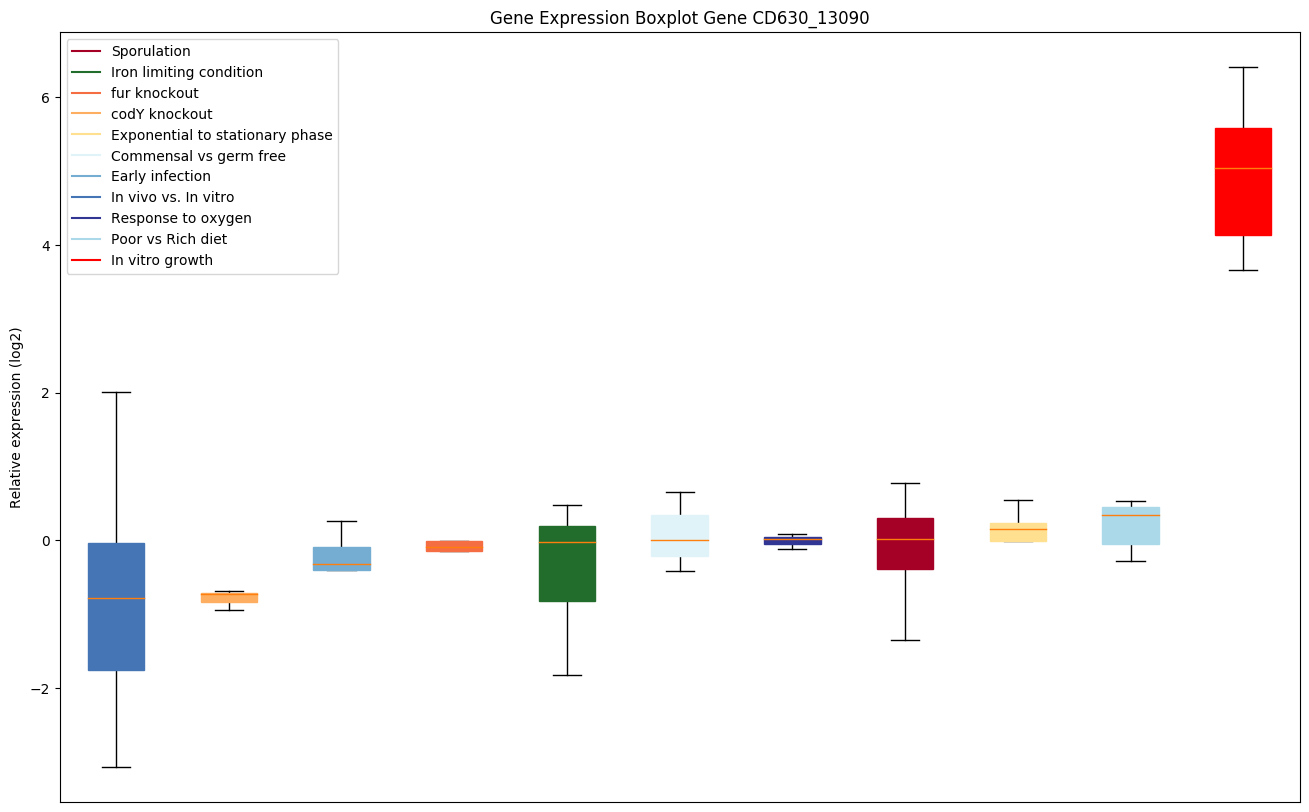 |
||
| CD630_07090 | mutS2 | DNA mismatch repair protein | Endonuclease that is involved in the suppression of homologous recombination and may therefore have a key role in the control of bacterial genetic diversity. |
 |
|||
| CD630_26820 | pfo | Pyruvate-ferredoxin oxidoreductase |
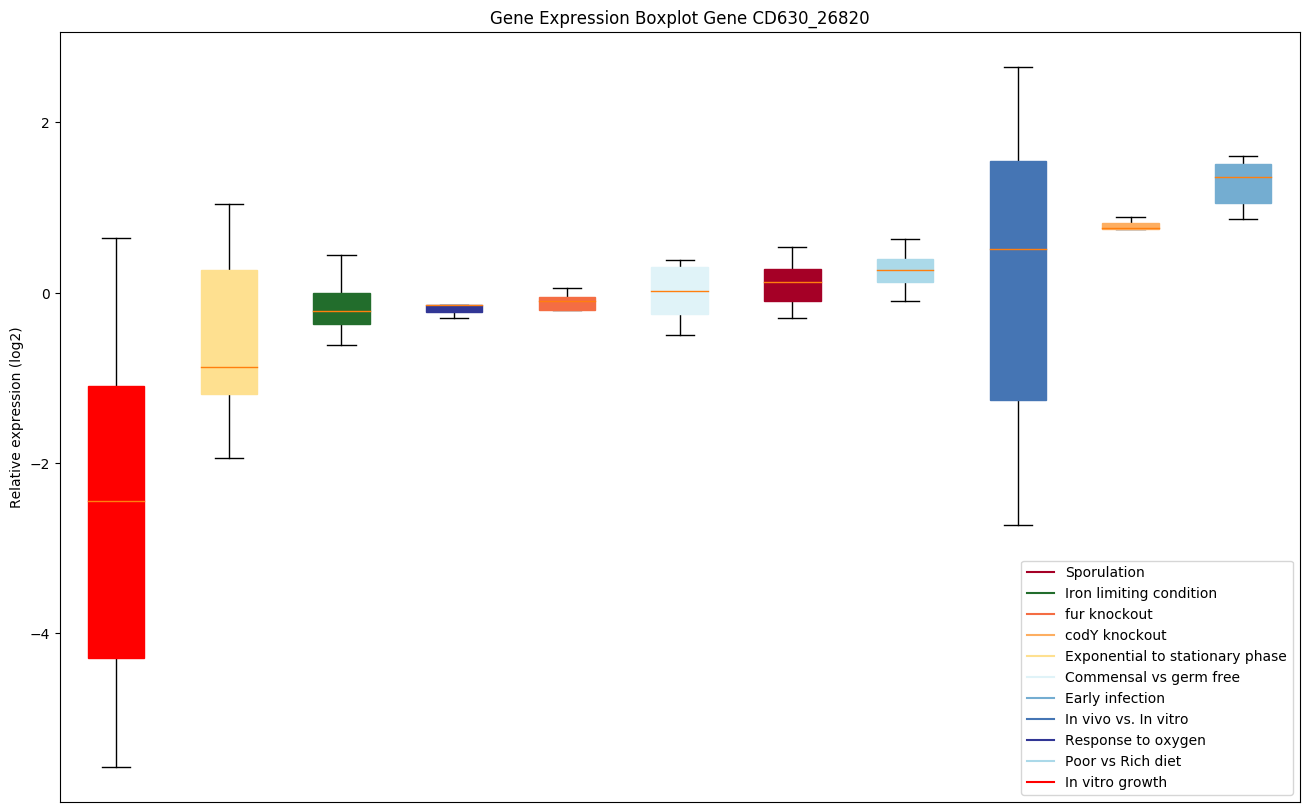 |
||||
| CD630_28030 | Putative membrane protein |
 |
|||||
| CD630_22740 | Conserved hypothetical protein |
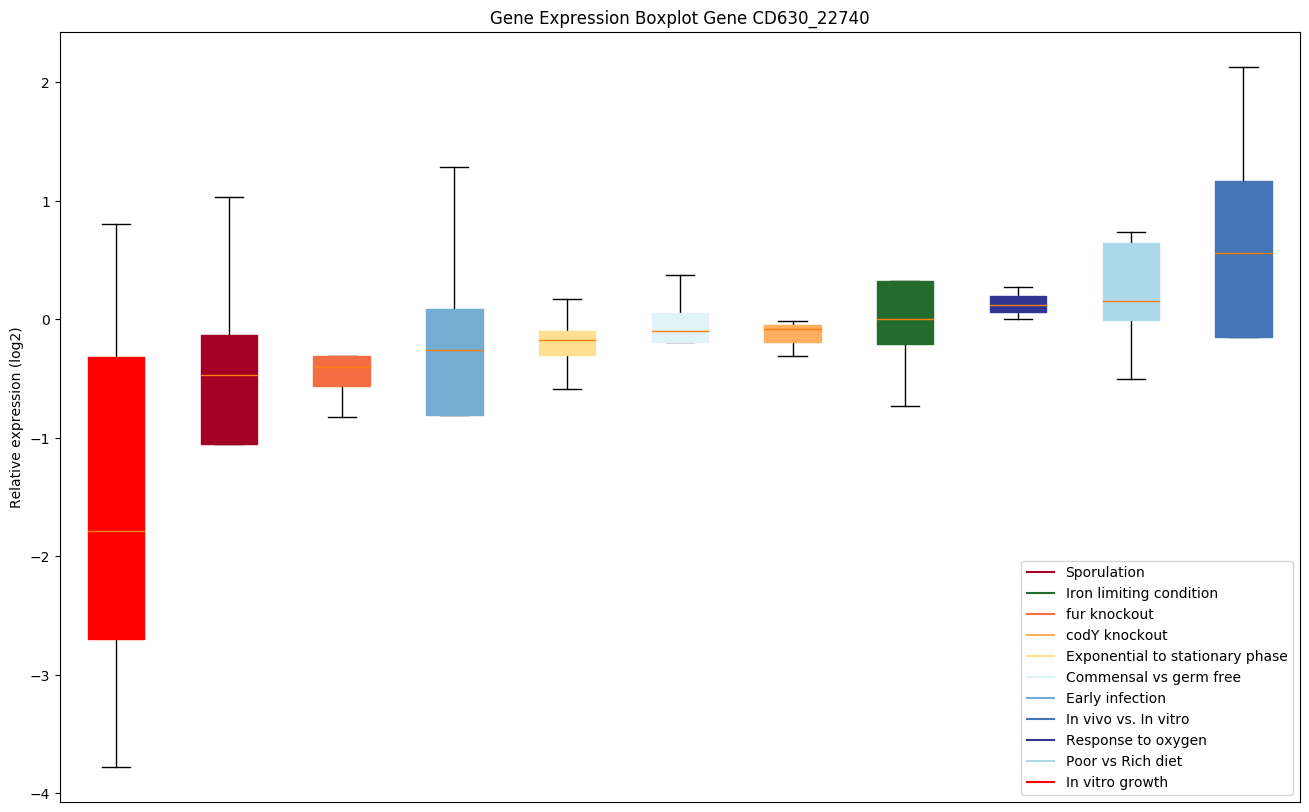 |
|||||
| CD630_28010 | Conserved hypothetical protein |
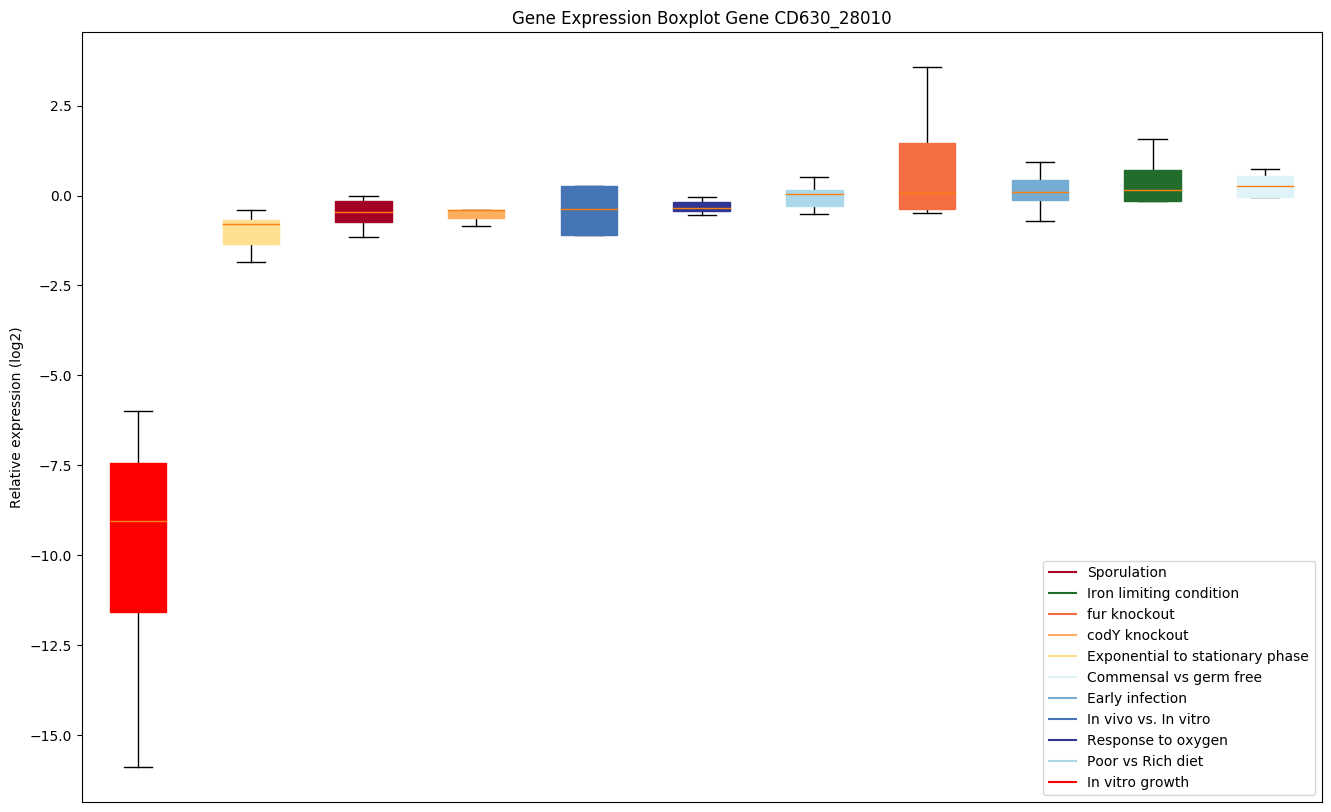 |
|||||
| CD630_13050 | dnaF | DNA polymerase III PolC-type (PolIII) | Required for replicative DNA synthesis. This DNA polymerase also exhibits 3' to 5' exonuclease activity. | Yes |
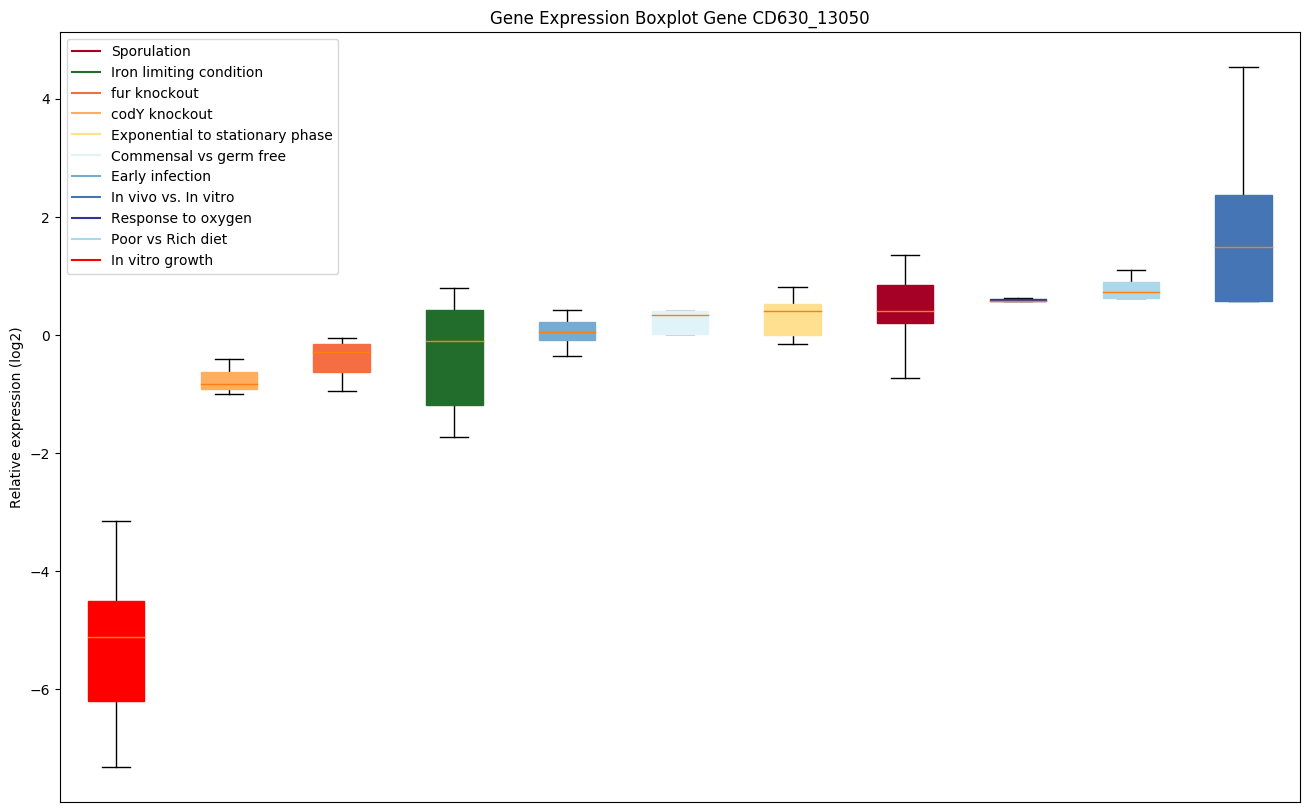 |