Module 211
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.52 | 23 | 63 | In vivo vs. In vitro | NA |
Bicluster expression profile
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
| TF | Module | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
|
spo0A Stage 0 sporulation protein A |
211 | 10 | 0.00 | 0.00 |
The module is significantly enriched with genes associated to the indicated functional terms
The enrichment was determined with hypergeometric test using the genome functional annotation compiled by Girinathan et al (2020).
| Module | Pathway | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
| 211 | DNA Recombination and Repair Systems | 5 | 0.000000 | 0.000133 |
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
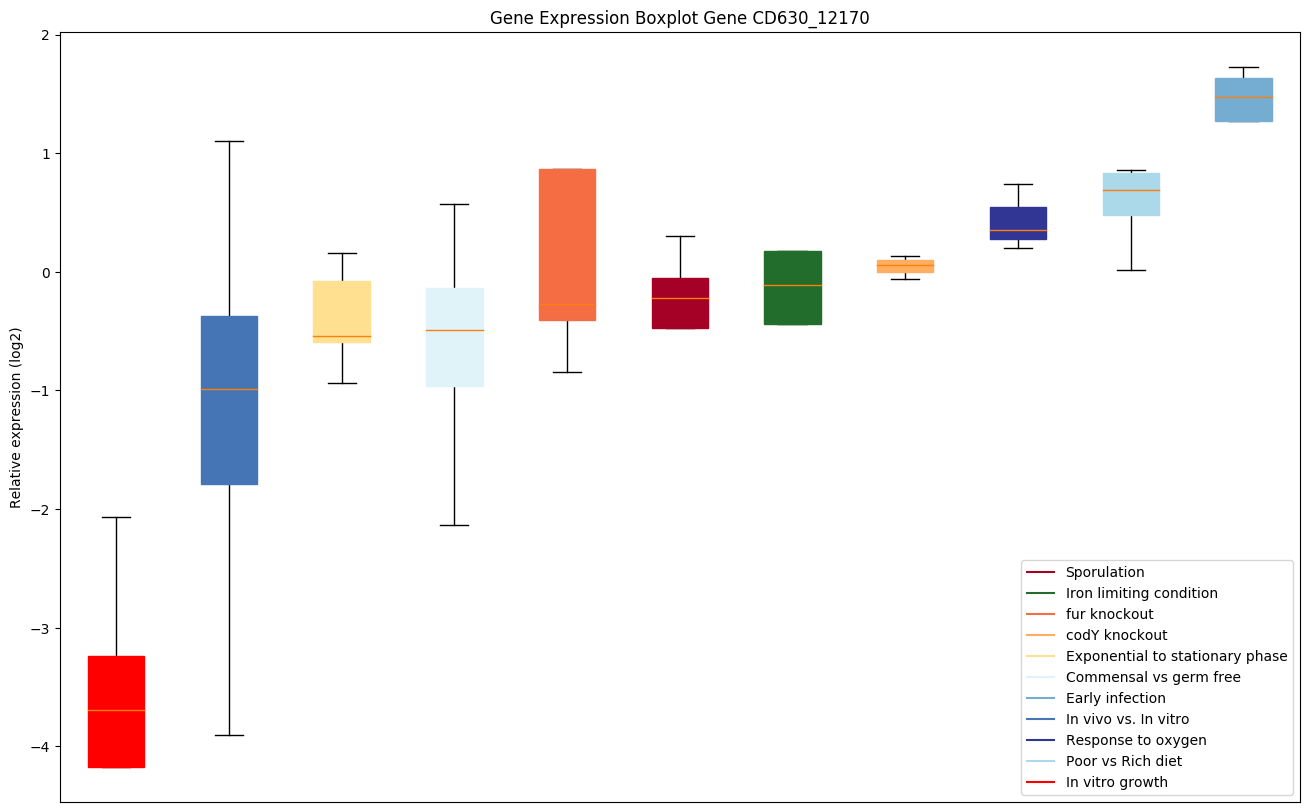
| CD630_12170 | Putative glycosyl transferase, family 2 | Yes |
 |
||||
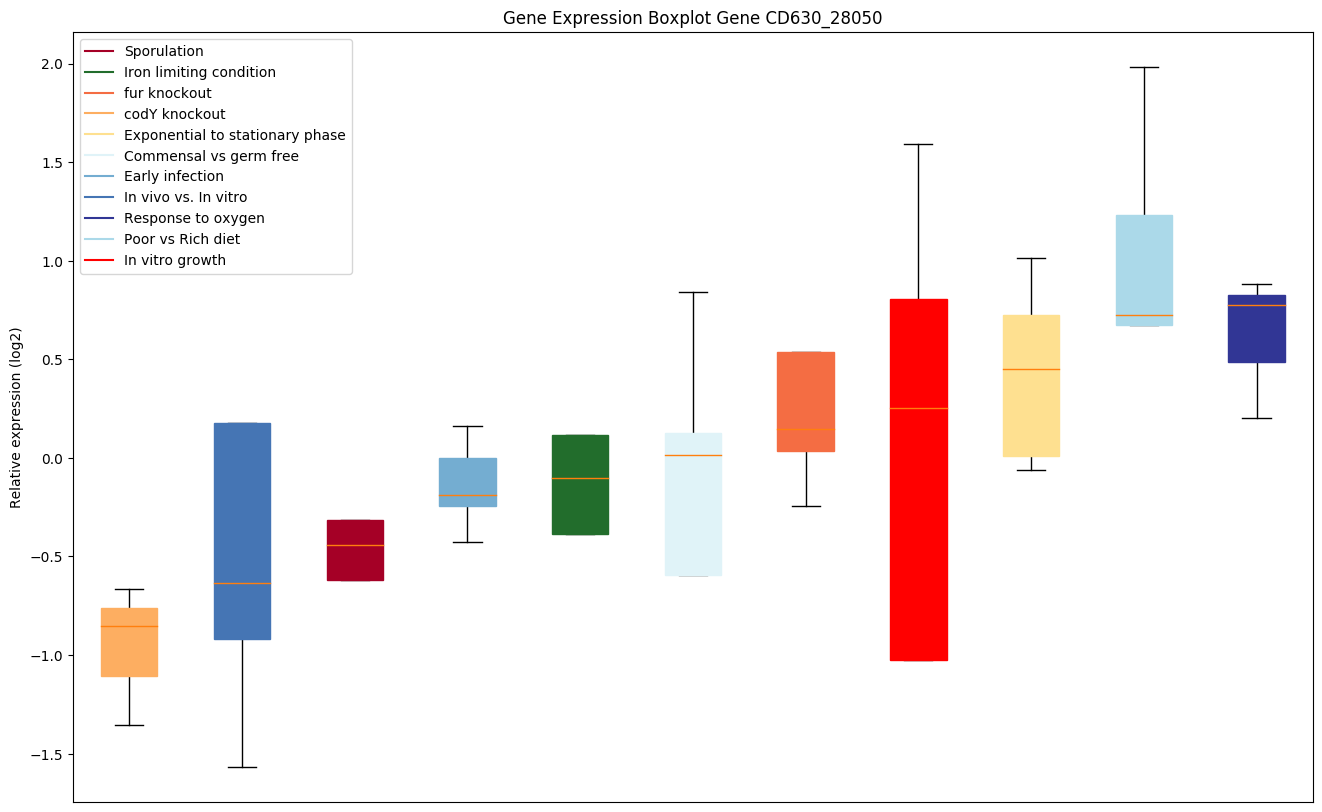
| CD630_28050 | ruvB | Holliday junction ATP-dependent DNA helicaseRuvB | The RuvA-RuvB complex in the presence of ATP renatures cruciform structure in supercoiled DNA with palindromic sequence, indicating that it may promote strand exchange reactions in homologous recombination. RuvAB is a helicase that mediates the Holliday junction migration by localized denaturation and reannealing. | Yes |
 |
||
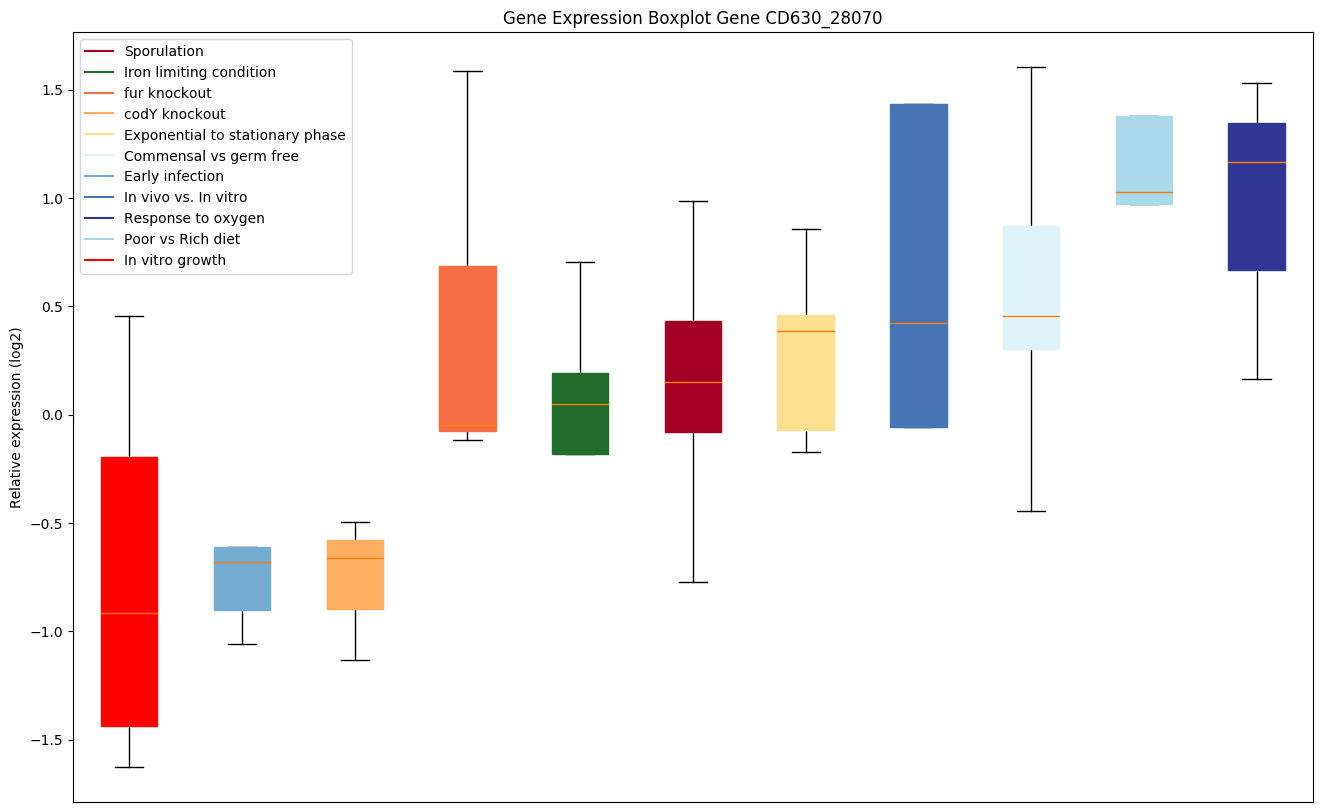
| CD630_28070 | ruvC | Crossover junction endodeoxyribonuclease RuvC(Holliday junction nuclease RuvC) (Holliday junctionresolvase RuvC) | Nuclease that resolves Holliday junction intermediates in genetic recombination. Cleaves the cruciform structure in supercoiled DNA by nicking to strands with the same polarity at sites symmetrically opposed at the junction in the homologous arms and leaves a 5'-terminal phosphate and a 3'-terminal hydroxyl group. |
 |
|||
| CD630_12150 | Putative pyrophosphokinase, DUF115 family | Yes |
 |
||||
| CD630_12230 | drm | Phosphopentomutase (Phosphodeoxyribomutase) | Phosphotransfer between the C1 and C5 carbon atoms of pentose. | Yes |
 |
||
| CD630_12250 | pdp | Pyrimidine-nucleoside phosphorylase | Yes | Yes |
 |
||
| CD630_12210 | Putative membrane protein |
 |
|||||
| CD630_12190 | Conserved hypothetical protein | Yes |
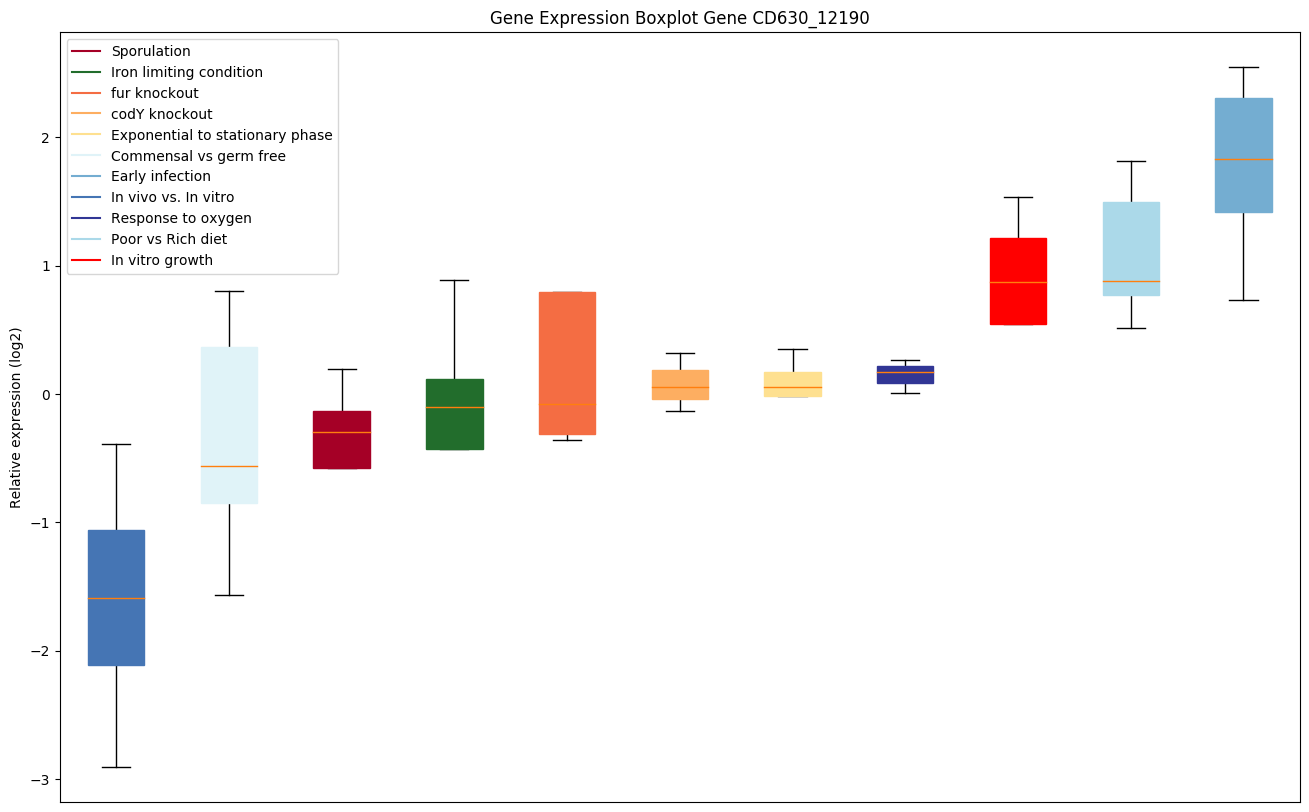 |
||||
| CD630_12310 | Putative site-specific recombinase |
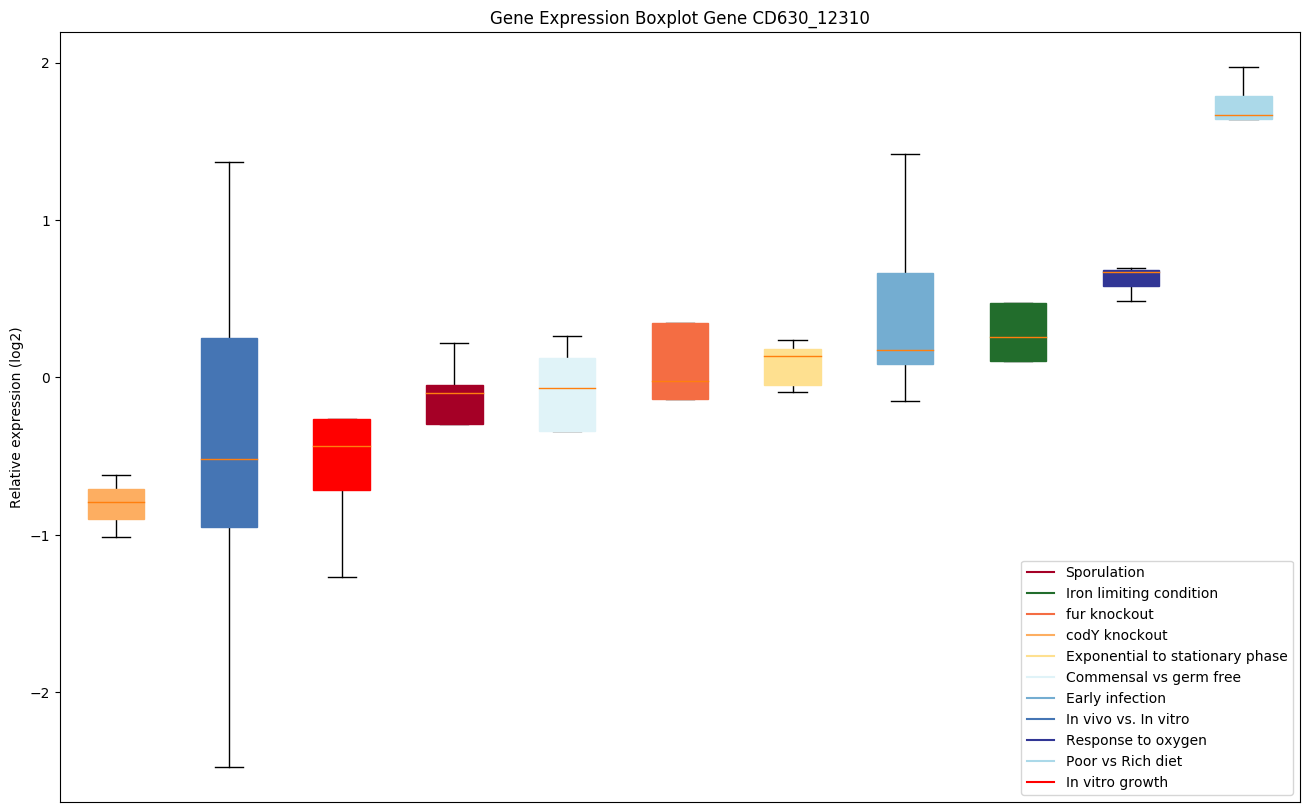 |
|||||
| CD630_22040 | Putative esterase |
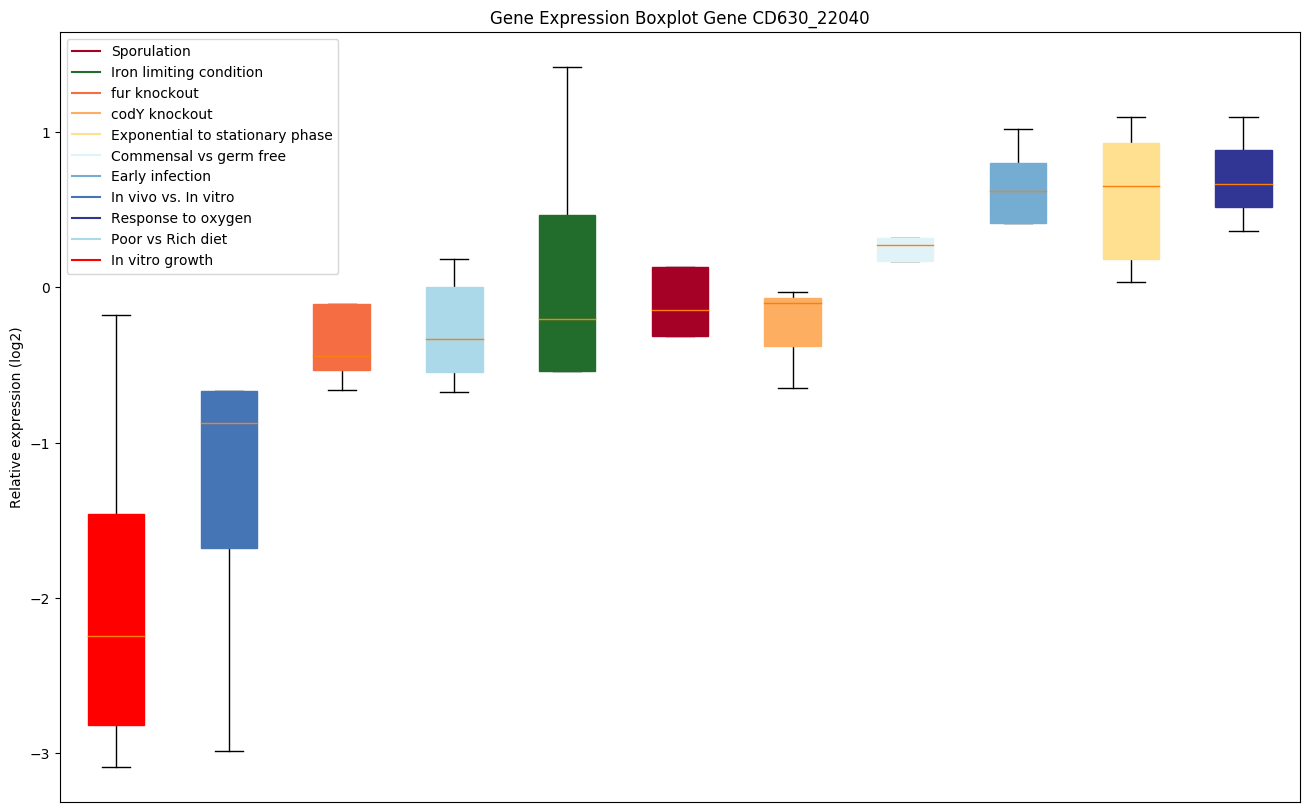 |
|||||
| CD630_22060 | aldH | Aldehyde dehydrogenase (aldehyde:NAD+oxidoreductase) |
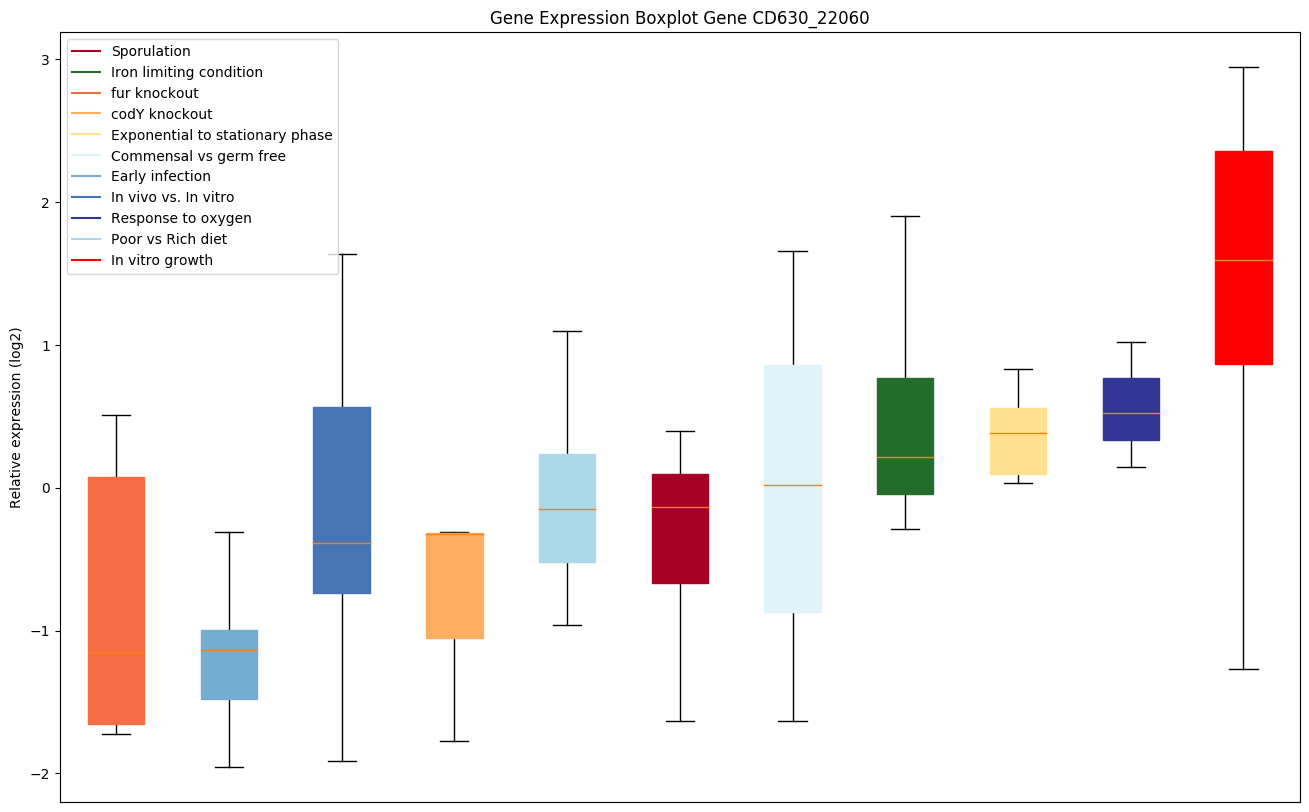 |
||||
| CD630_28060 | ruvA | Holliday junction ATP-dependent DNA helicaseRuvA | The RuvA-RuvB complex in the presence of ATP renatures cruciform structure in supercoiled DNA with palindromic sequence, indicating that it may promote strand exchange reactions in homologous recombination. RuvAB is a helicase that mediates the Holliday junction migration by localized denaturation and reannealing. RuvA stimulates, in the presence of DNA, the weak ATPase activity of RuvB. |
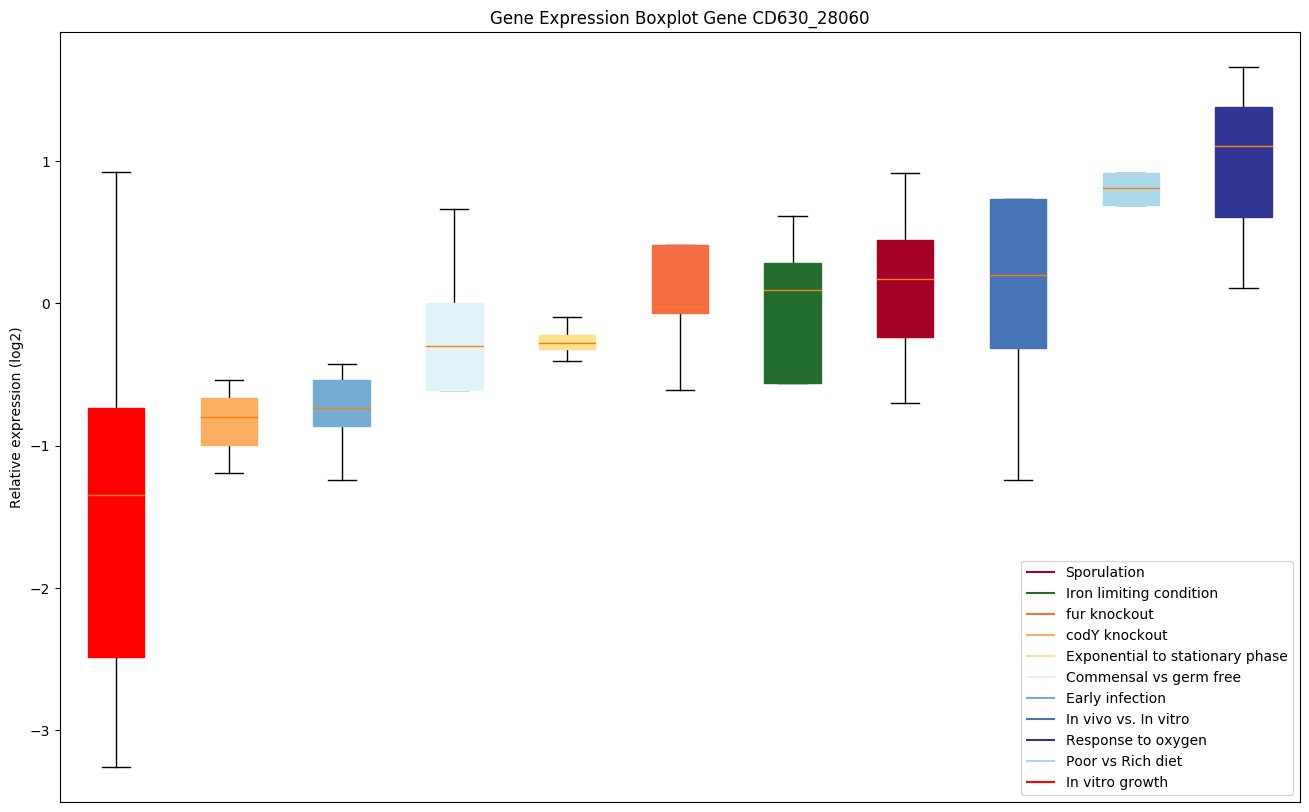 |
|||
| CD630_26460 | ftsZ | Cell division protein FtsZ | Essential cell division protein that forms a contractile ring structure (Z ring) at the future cell division site. The regulation of the ring assembly controls the timing and the location of cell division. One of the functions of the FtsZ ring is to recruit other cell division proteins to the septum to produce a new cell wall between the dividing cells. Binds GTP and shows GTPase activity. | Yes |
 |
||
| CD630_12160 | Conserved hypothetical protein | Yes |
 |
||||
| CD630_14010 | Putative isochorismatase |
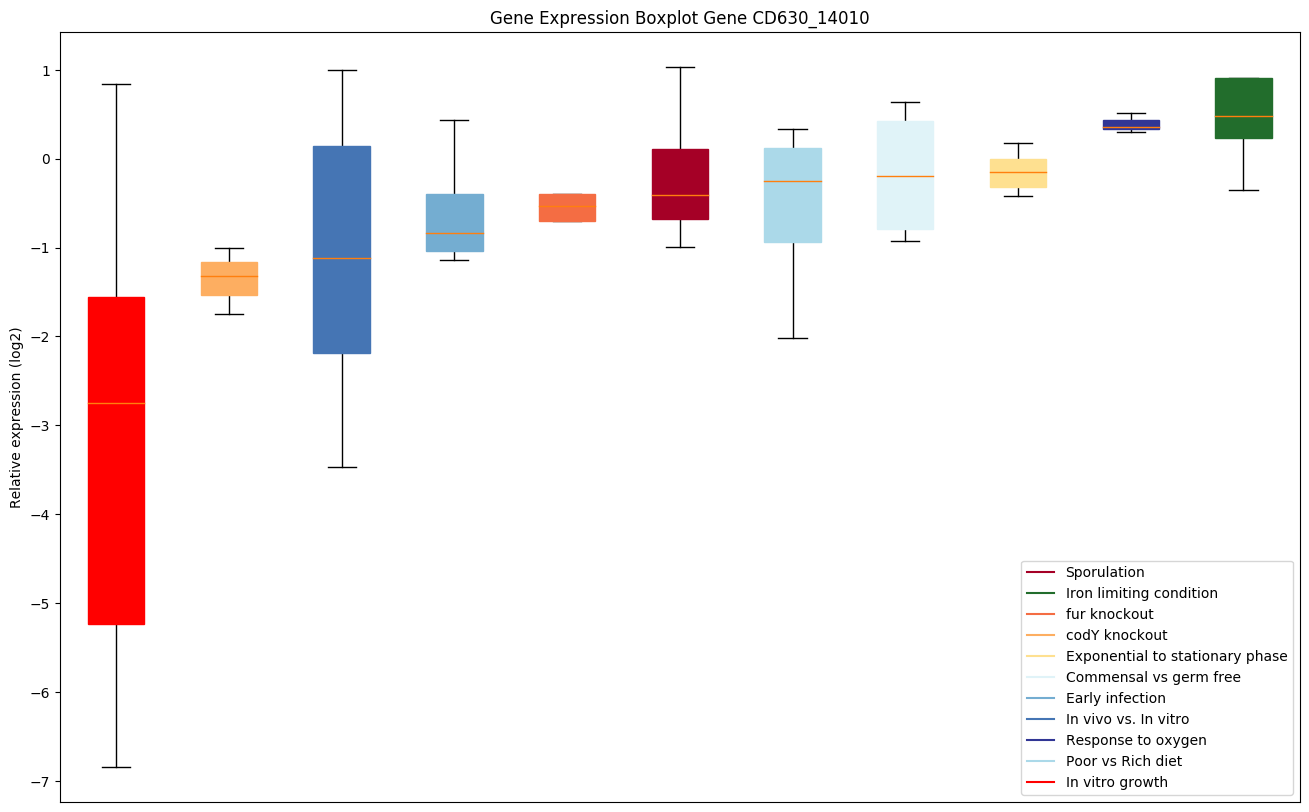 |
|||||
| CD630_12300 | RNA polymerase sigma-K factor SigK (Part 2) | Sigma factors are initiation factors that promote the attachment of RNA polymerase to specific initiation sites and are then released. |
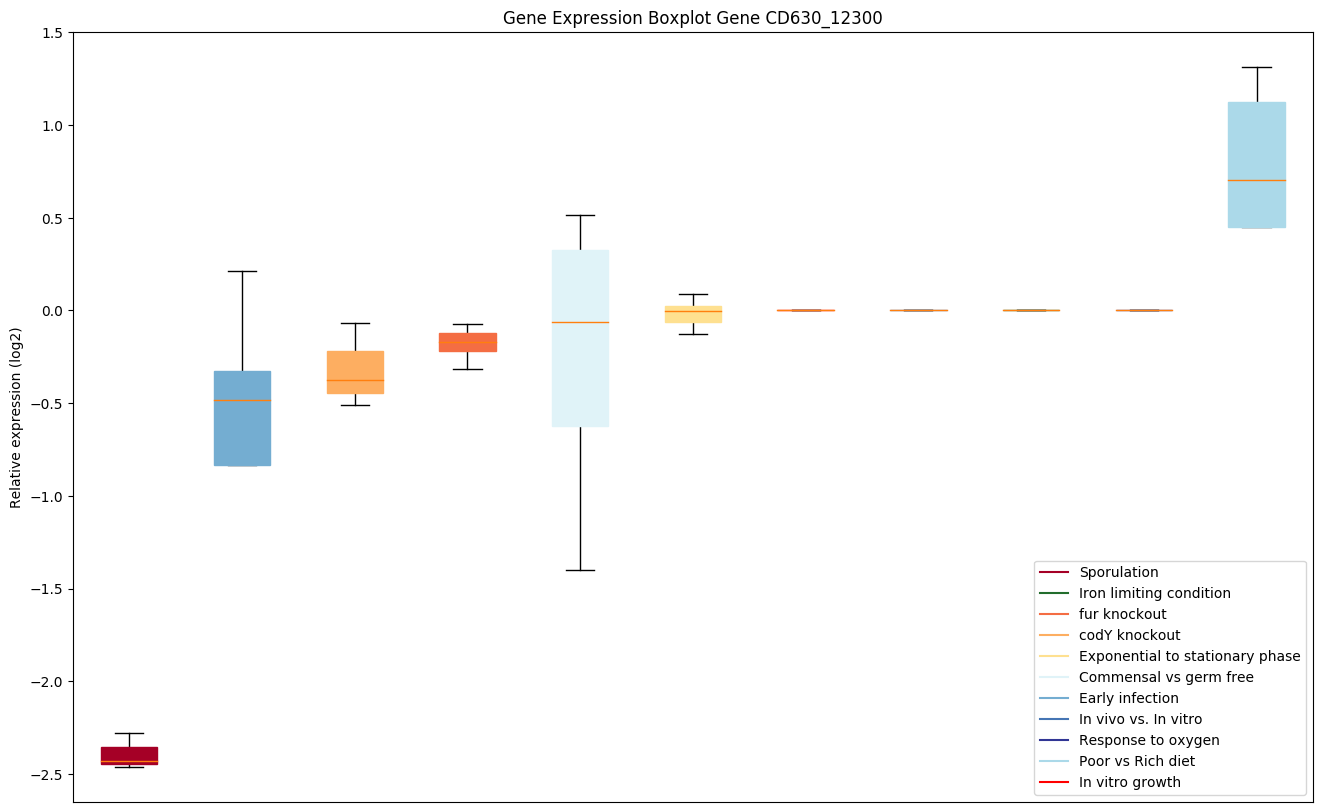 |
||||
| CD630_12240 | pupG | Purine nucleoside phosphorylase | The purine nucleoside phosphorylases catalyze the phosphorolytic breakdown of the N-glycosidic bond in the beta-(deoxy)ribonucleoside molecules, with the formation of the corresponding free purine bases and pentose-1-phosphate. | Yes |
 |
||
| CD630_12180 | Putative glycosyl transferase | Yes |
 |
||||
| CD630_12200 | nudF | Putative hydrolase, NUDIX family | Yes |
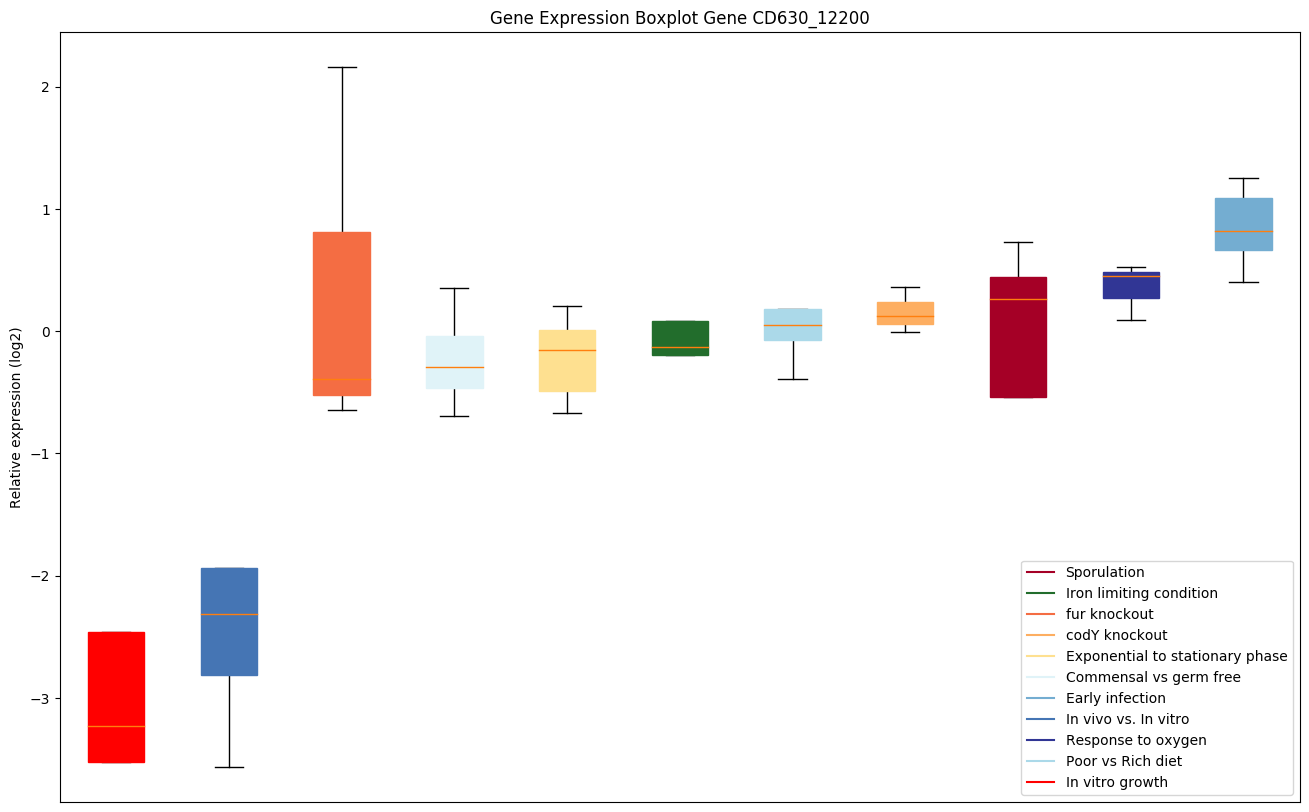 |
|||
| CD630_12220 | Putative phage integrase site-specificrecombinase XerD-like |
 |
|||||
| CD630_22050 | Putative membrane protein |
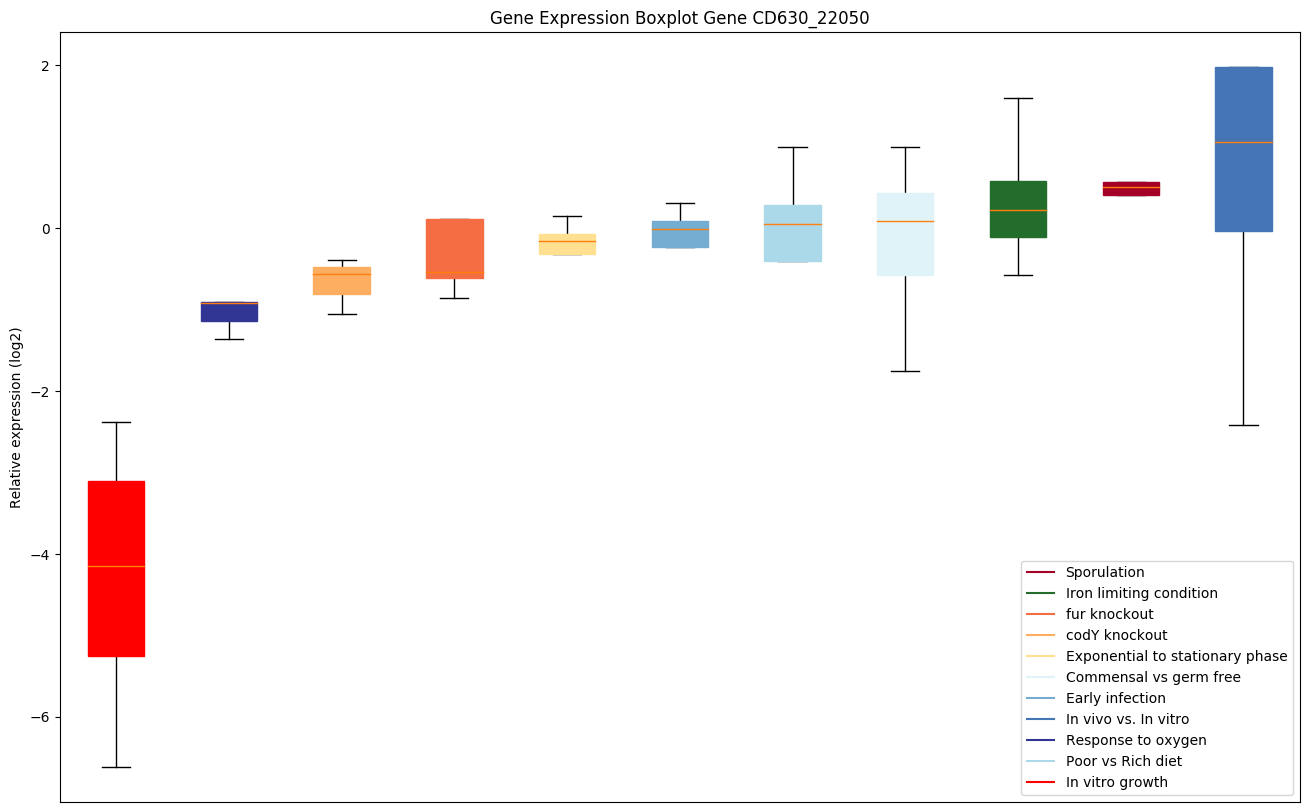 |
|||||
| CD630_22290 | Putative membrane protein |
 |
|||||
| CD630_14020 | glpQ | Glycerophosphoryl diester phosphodiesterase |
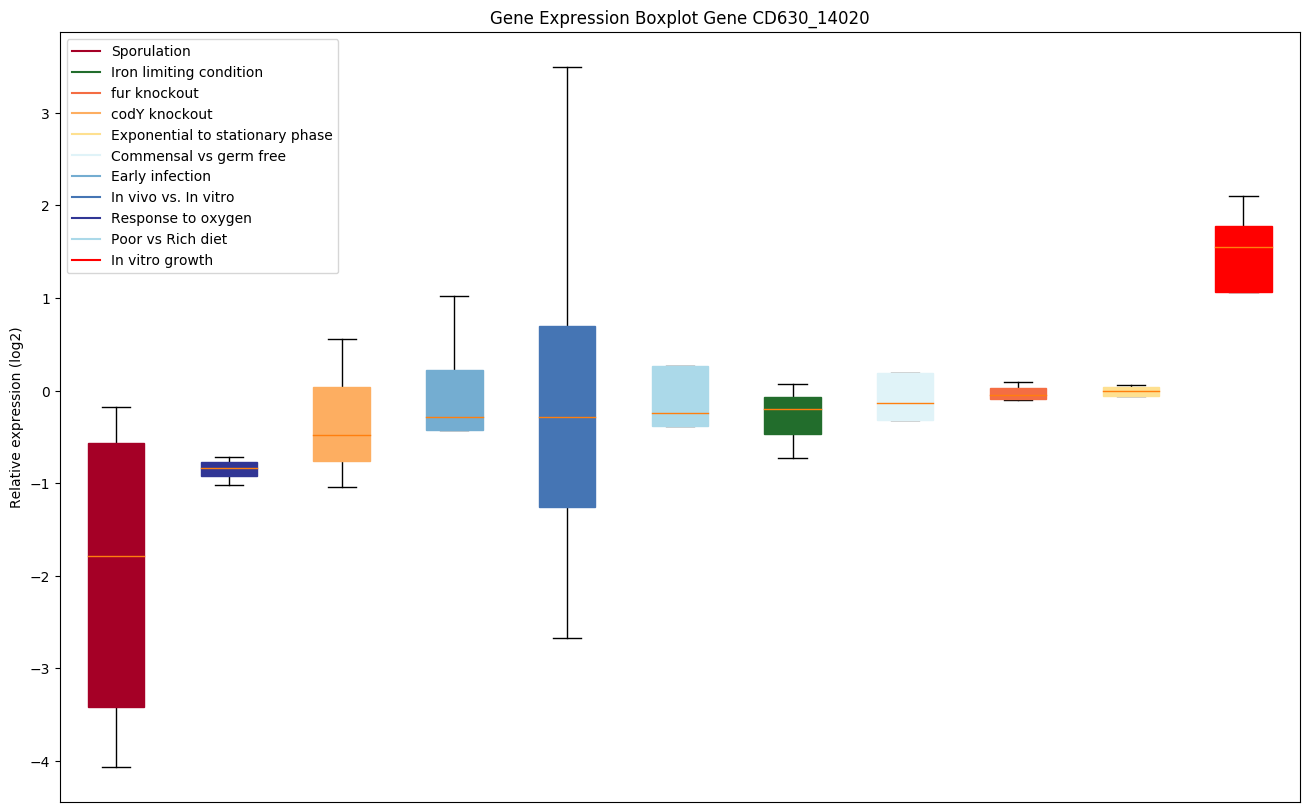 |