Module 368
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.54 | 28 | 65 | NA | NA |
Bicluster expression profile
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
| TF | Module | Confidence Score | Beta |
|---|---|---|---|
|
fapR Transcriptional regulator, DeoR family (Fattyacid and phospholipid biosynthesis regulator) |
368 | 0.54 | 0.2260 |
The module is significantly enriched with genes associated to the indicated functional terms
The enrichment was determined with hypergeometric test using the genome functional annotation compiled by Girinathan et al (2020).
| Module | Pathway | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
| 368 | Mannose, Sorbose, Fructose Transport and Conversion | 6 | 0.000000 | 0.000003 |
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
| CD630_06980 | Putative tRNA/rRNA methyltransferase, SpoU |
 |
|||||
| CD630_10770 | PTS system, mannose/fructose/sorbose IICcomponent |
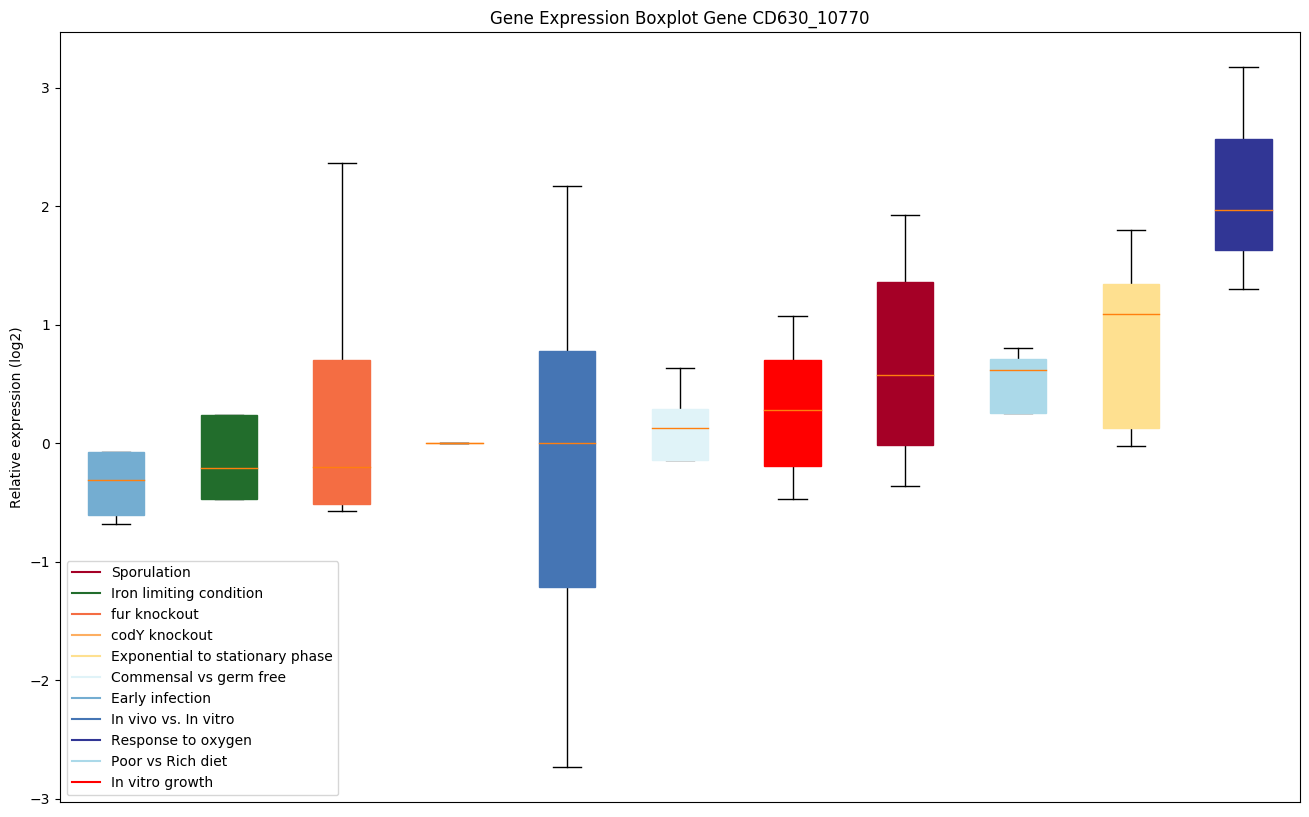 |
|||||
| CD630_17330 | hemE | Uroporphyrinogen decarboxylase (URO-D) |
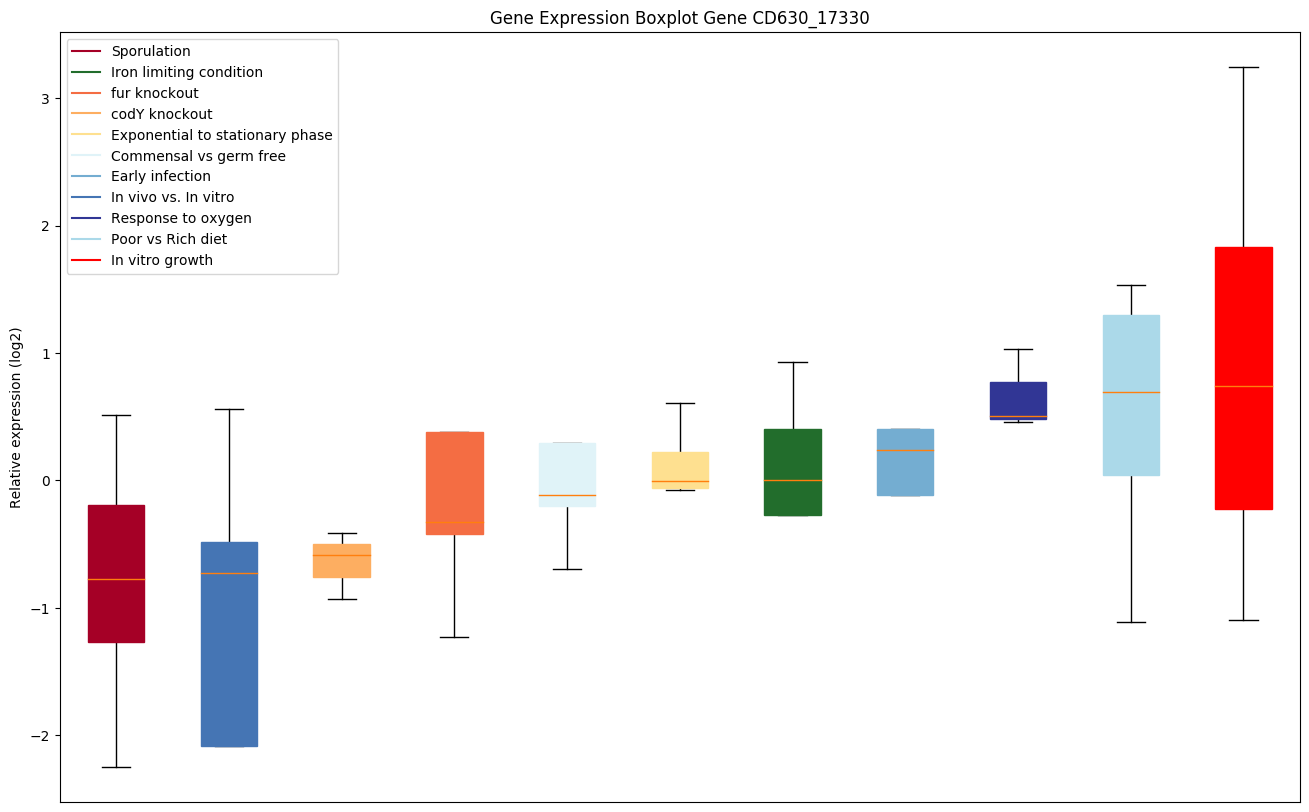 |
||||
| CD630_00070 | Conserved hypothetical protein |
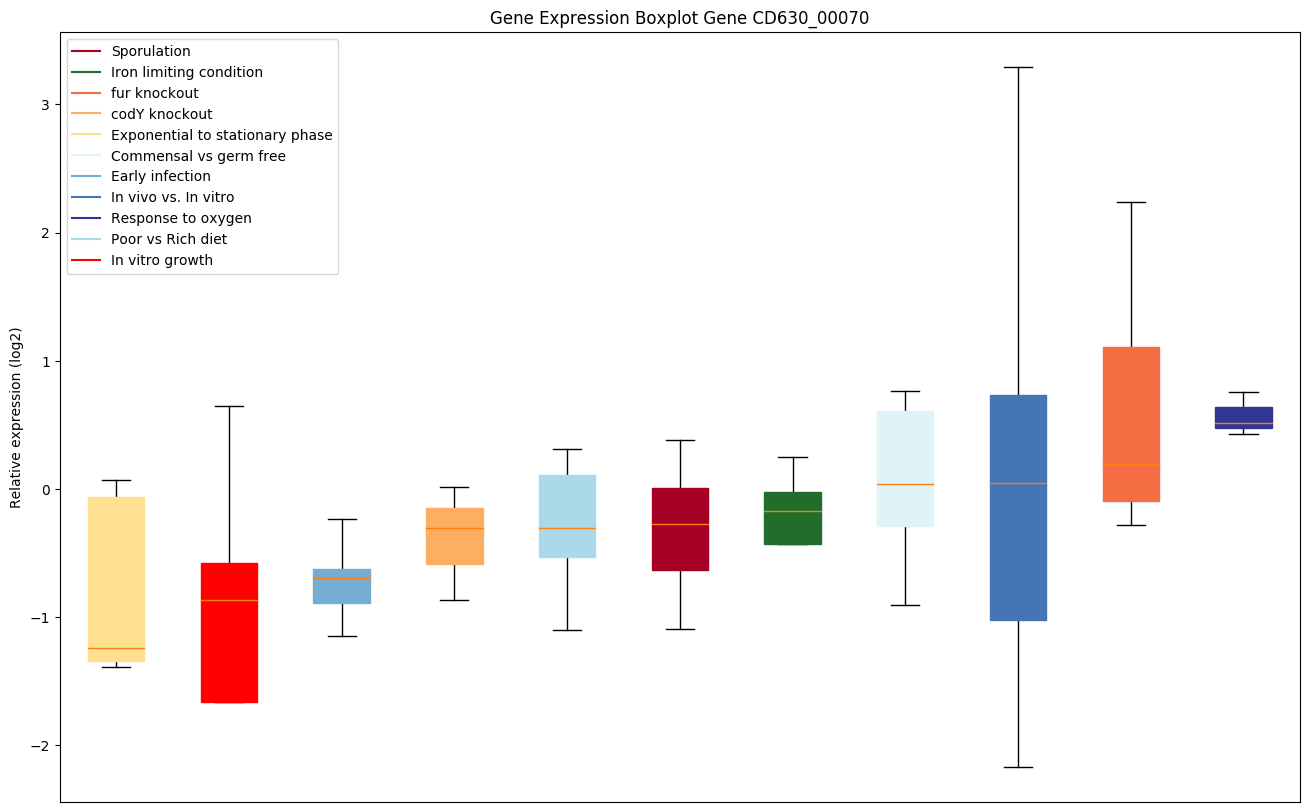 |
|||||
| CD630_00050 | gyrB | DNA gyrase subunit B | A type II topoisomerase that negatively supercoils closed circular double-stranded (ds) DNA in an ATP-dependent manner to modulate DNA topology and maintain chromosomes in an underwound state. Negative supercoiling favors strand separation, and DNA replication, transcription, recombination and repair, all of which involve strand separation. Also able to catalyze the interconversion of other topological isomers of dsDNA rings, including catenanes and knotted rings. Type II topoisomerases break and join 2 DNA strands simultaneously in an ATP-dependent manner. |
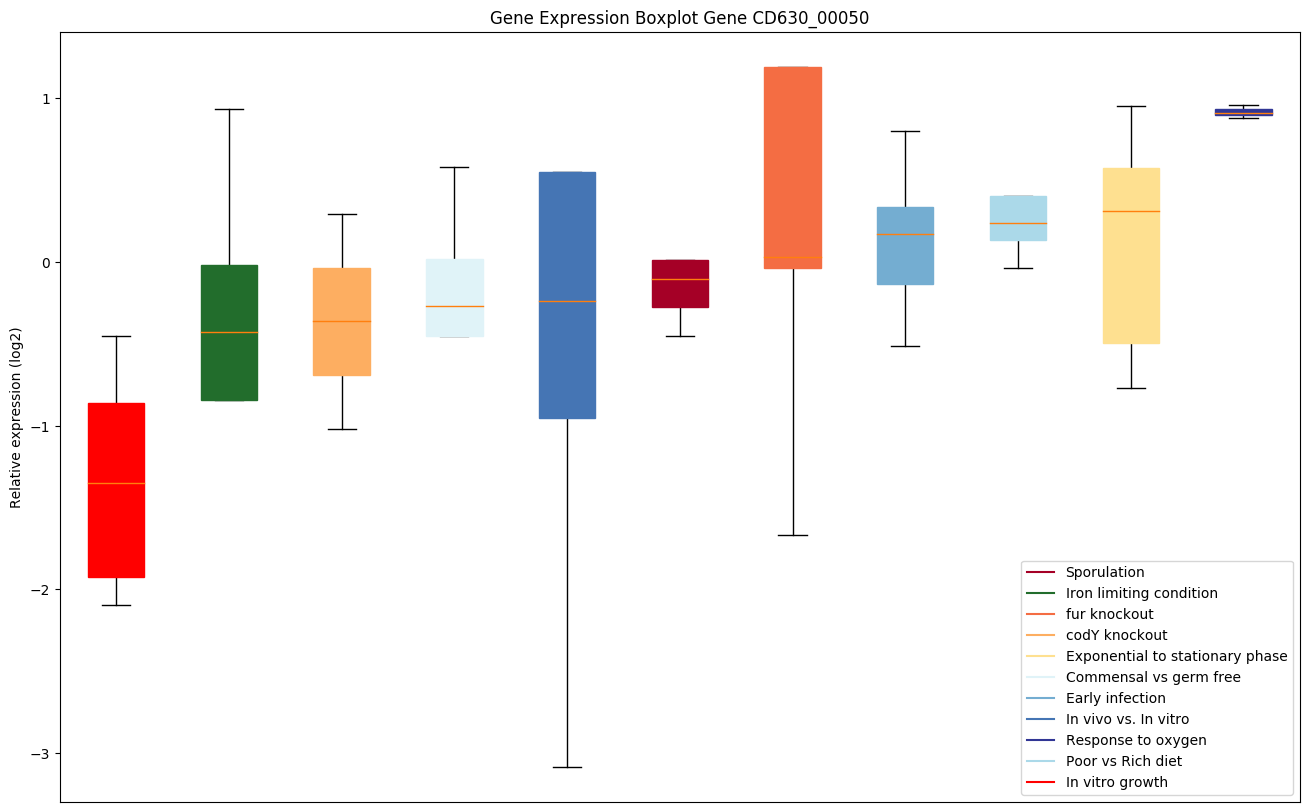 |
|||
| CD630_10750 | Conserved hypothetical protein |
 |
|||||
| CD630_00090 | rsbV | Anti-anti-sigma factor |
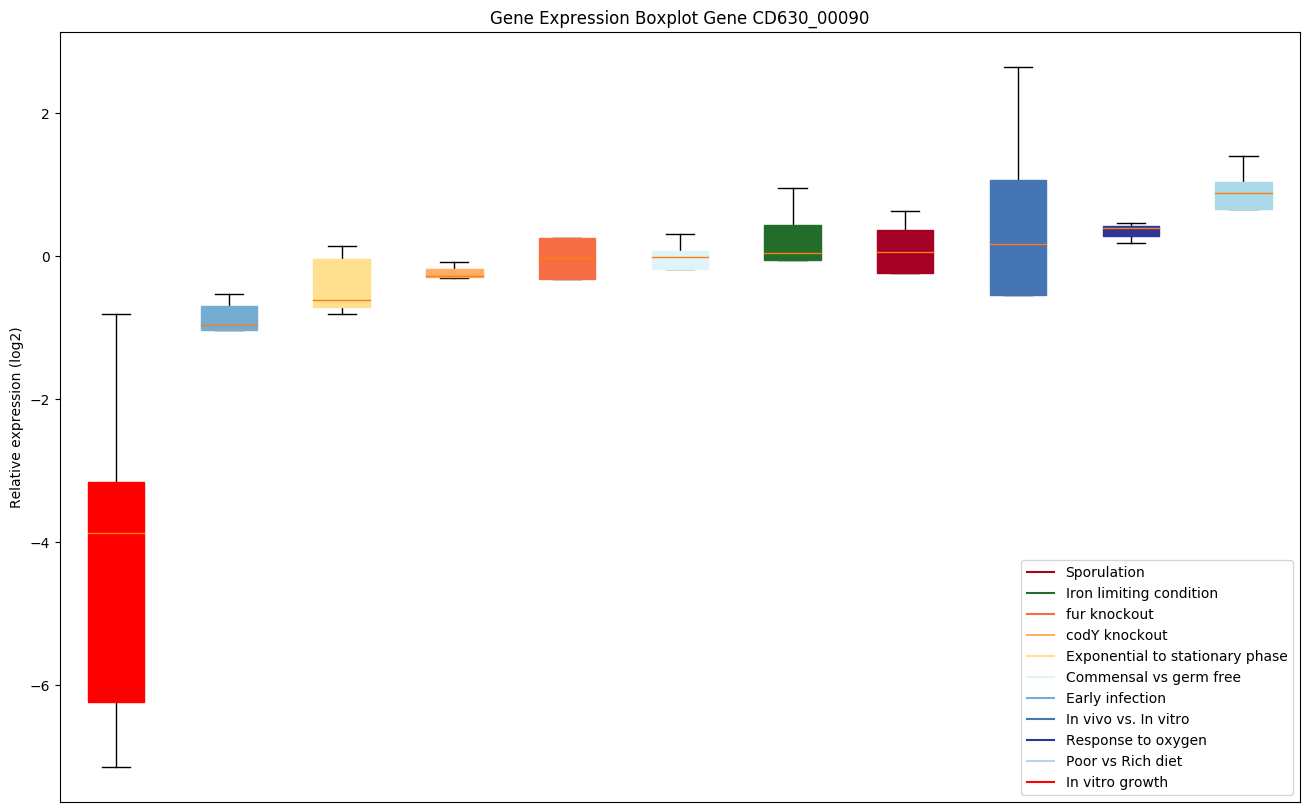 |
||||
| CD630_10730 | Transcriptional regulator, GntR family |
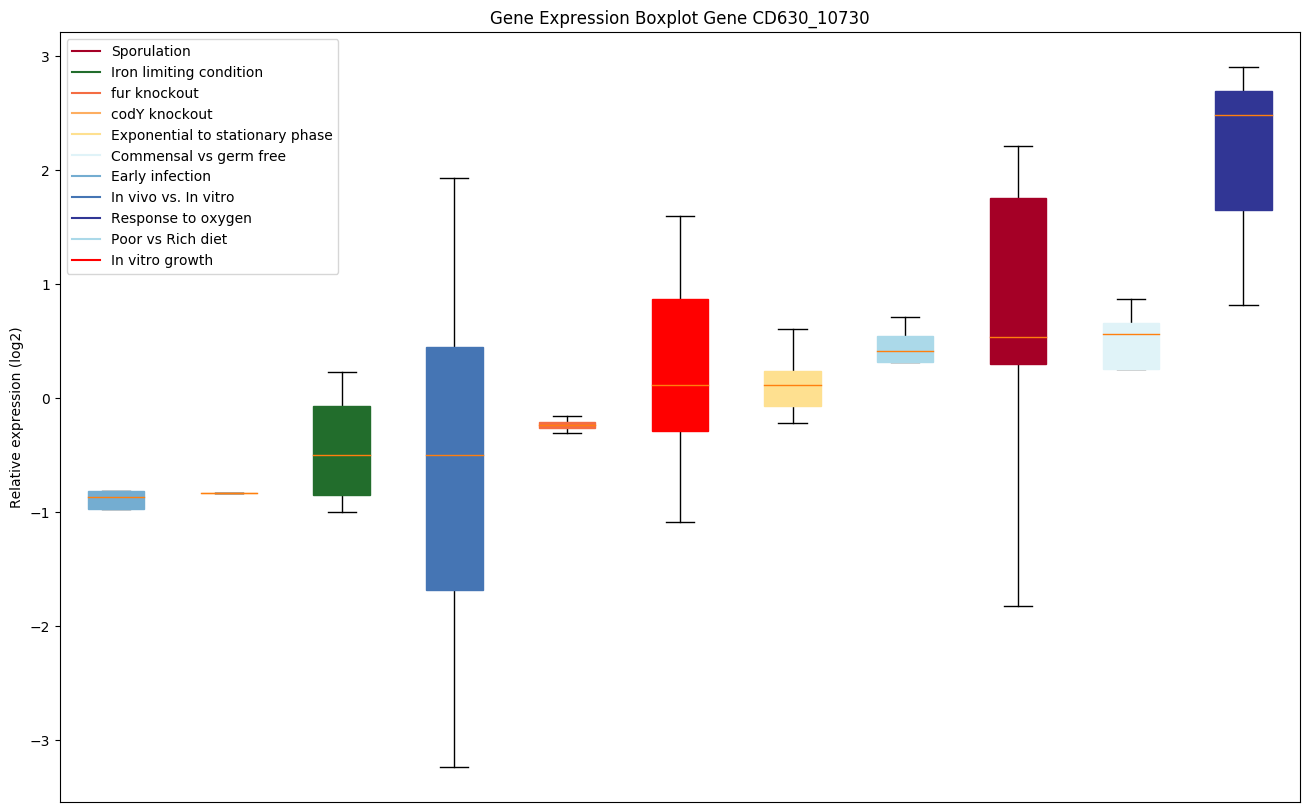 |
|||||
| CD630_14070 | Transcriptional regulator, GntR family | Yes |
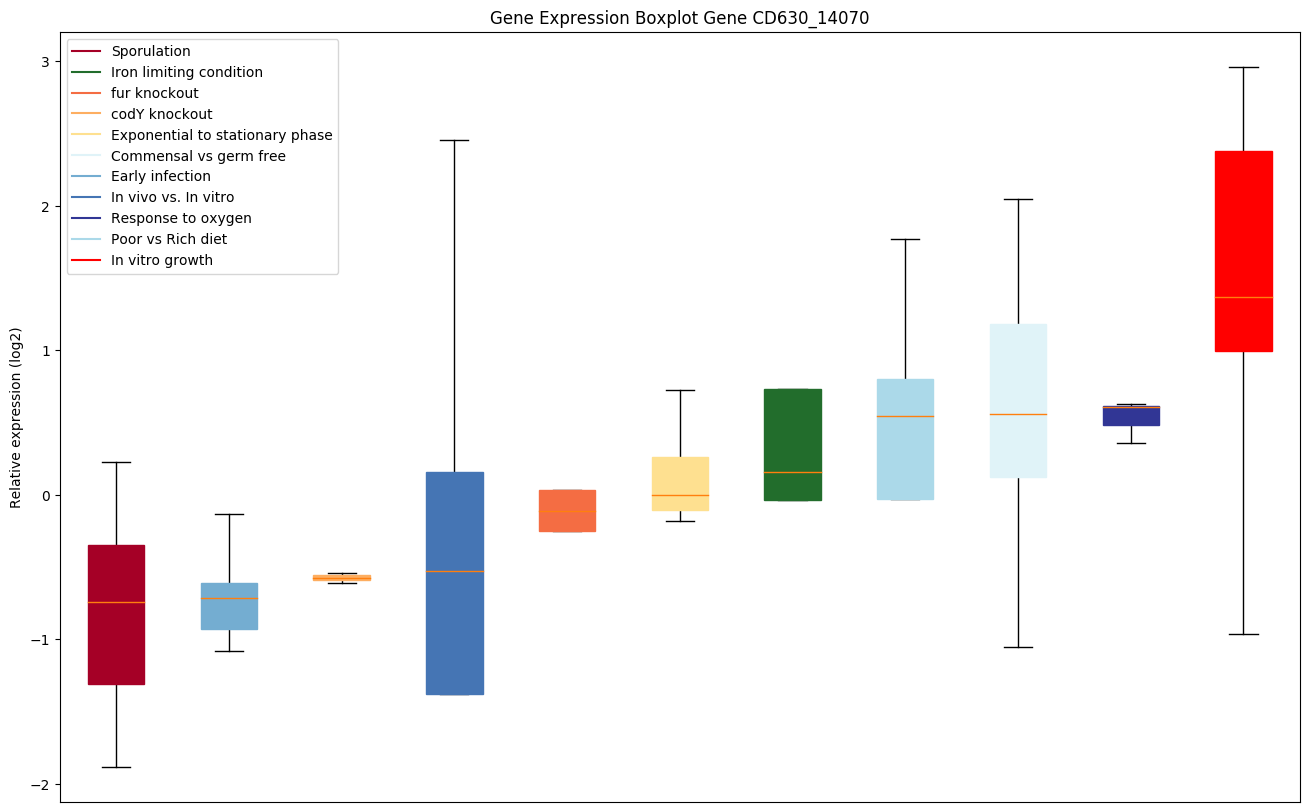 |
||||
| CD630_22640 | Conserved hypothetical protein |
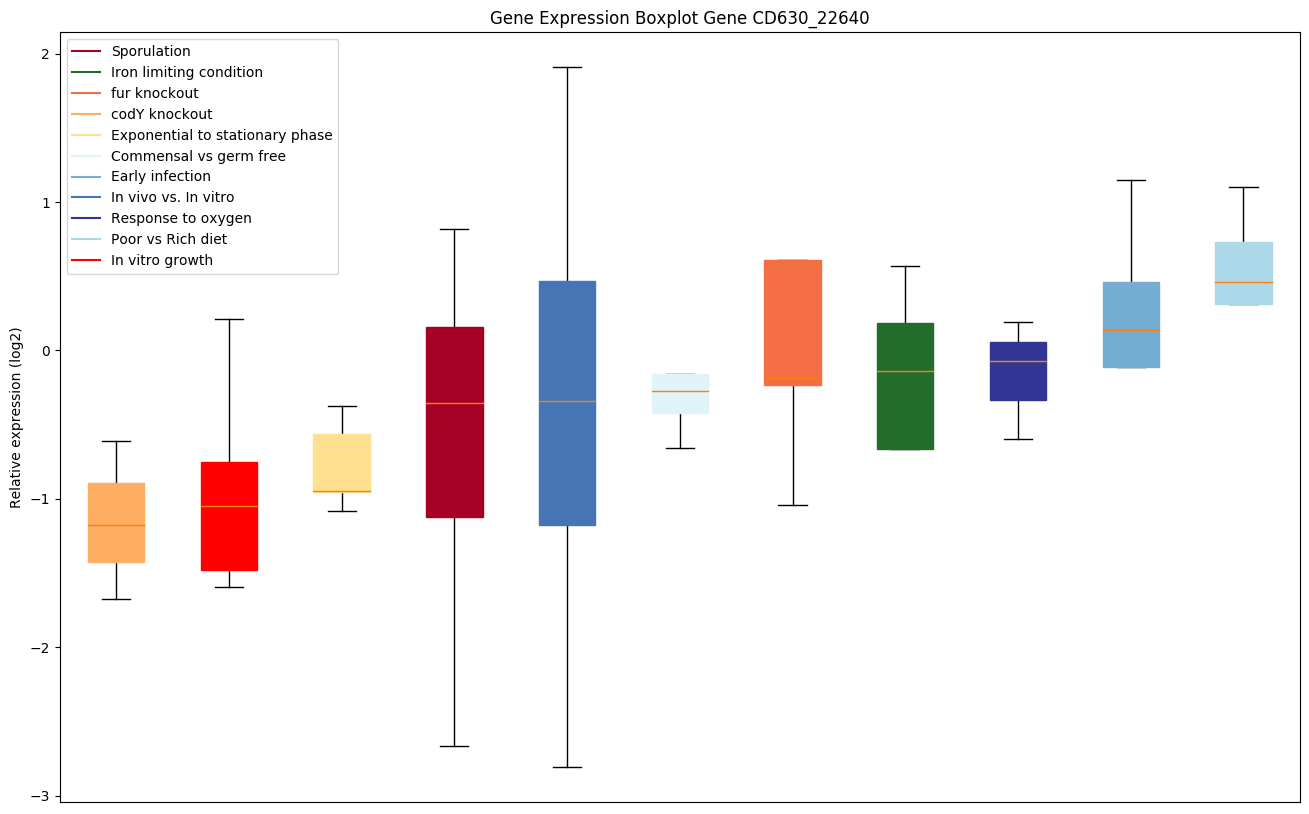 |
|||||
| CD630_14050 | ABC-type transport system, ATP-binding protein |
 |
|||||
| CD630_15300 | Two-component sensor histidine kinase |
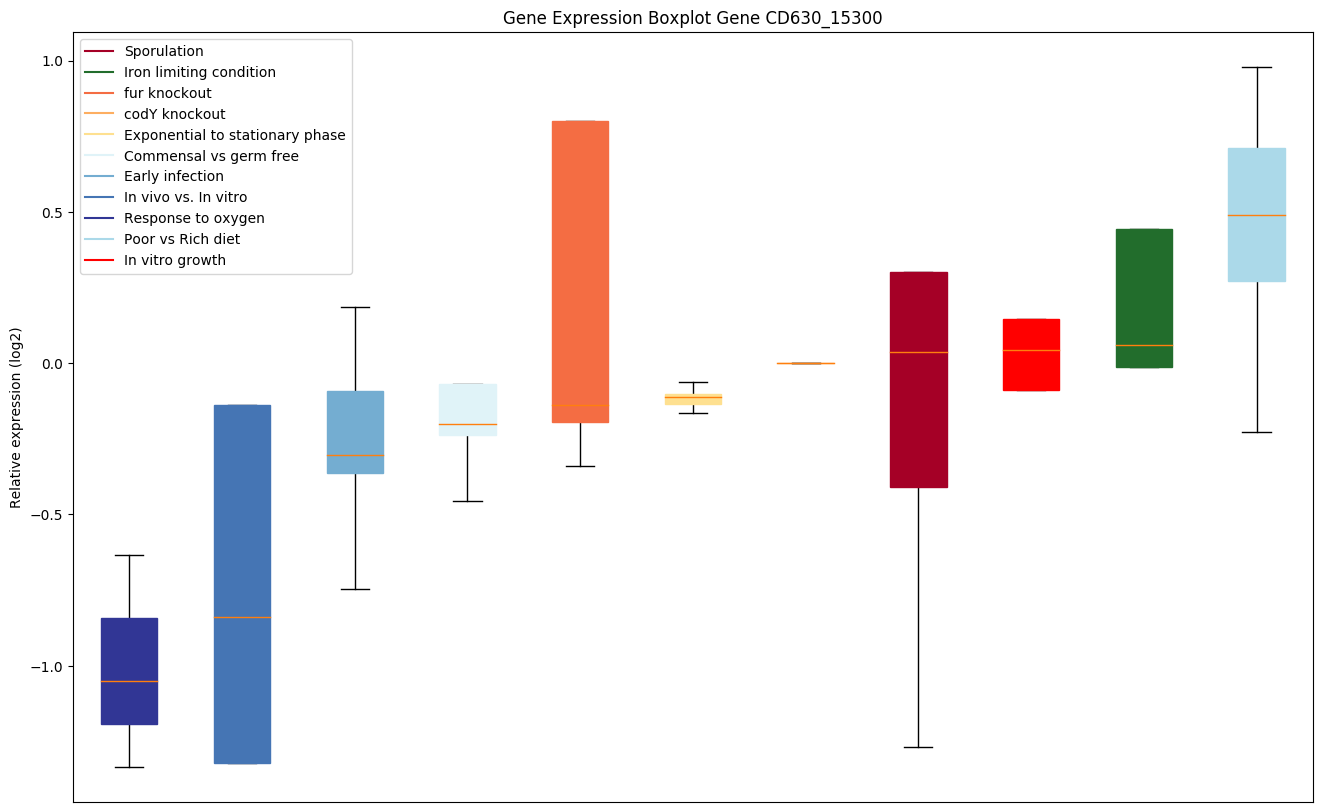 |
|||||
| CD630_10780 | PTS system, mannose/fructose/sorbose IIDcomponent |
 |
|||||
| CD630_00110 | sigB | RNA polymerase sigma-B factor |
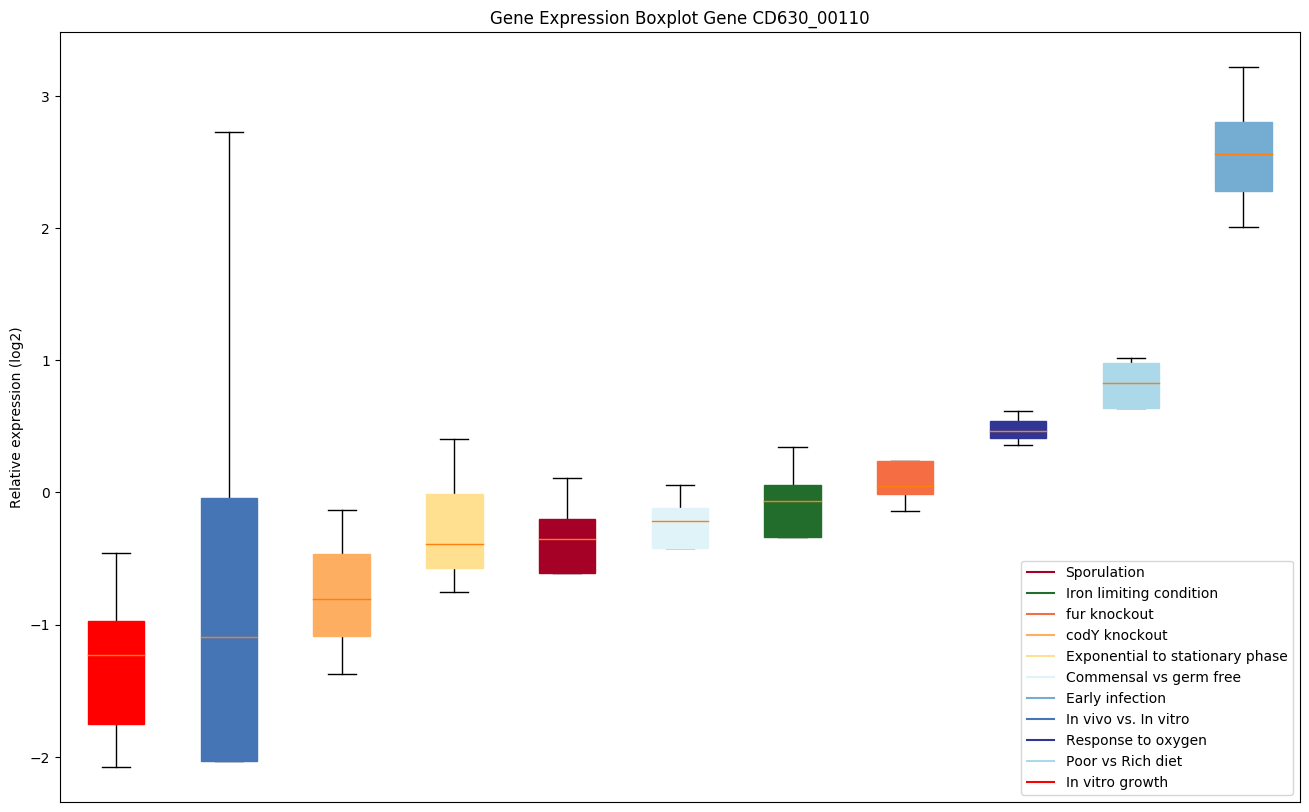 |
||||
| CD630_00040 | recF | DNA replication and repair protein RecF | The RecF protein is involved in DNA metabolism; it is required for DNA replication and normal SOS inducibility. RecF binds preferentially to single-stranded, linear DNA. It also seems to bind ATP. |
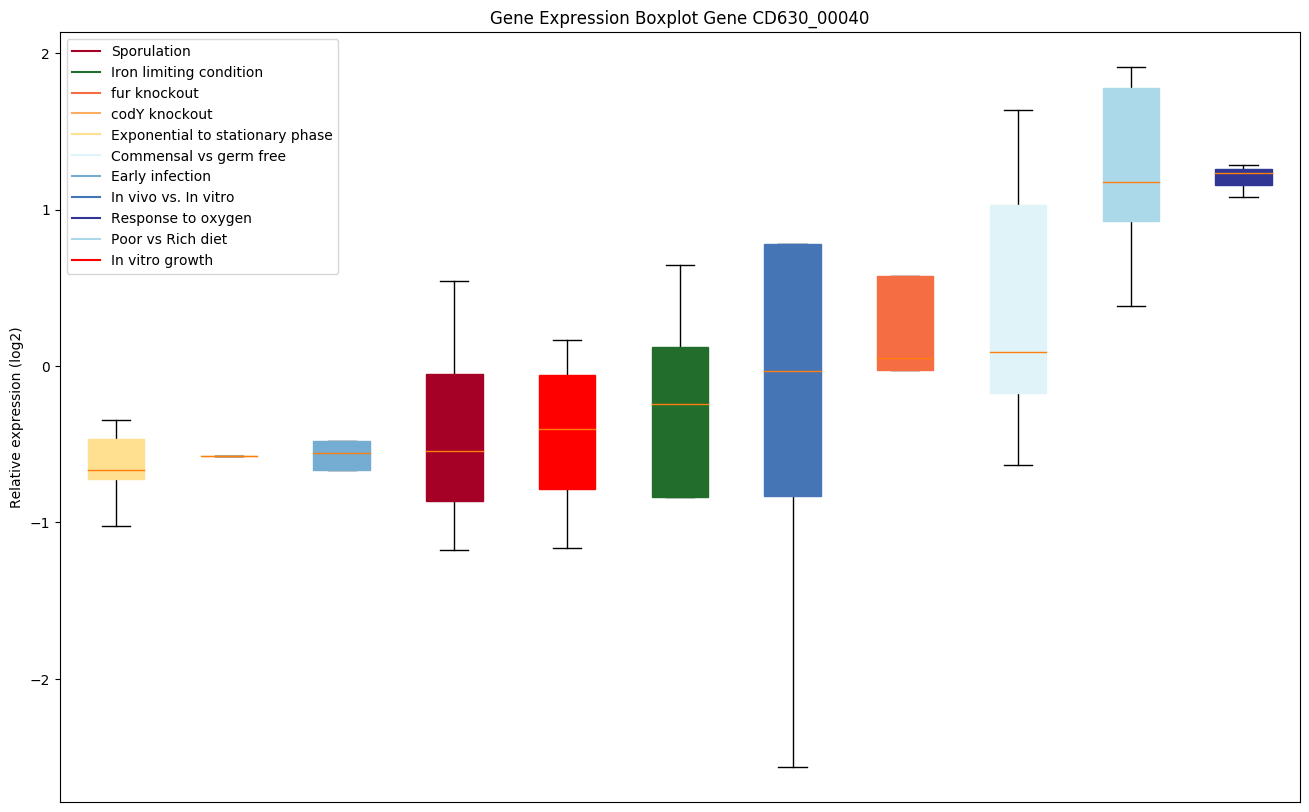 |
|||
| CD630_00020 | dnaN | DNA polymerase III subunit beta | Confers DNA tethering and processivity to DNA polymerases and other proteins. Acts as a clamp, forming a ring around DNA (a reaction catalyzed by the clamp-loading complex) which diffuses in an ATP-independent manner freely and bidirectionally along dsDNA. Initially characterized for its ability to contact the catalytic subunit of DNA polymerase III (Pol III), a complex, multichain enzyme responsible for most of the replicative synthesis in bacteria; Pol III exhibits 3'-5' exonuclease proofreading activity. The beta chain is required for initiation of replication as well as for processivity of DNA replication. |
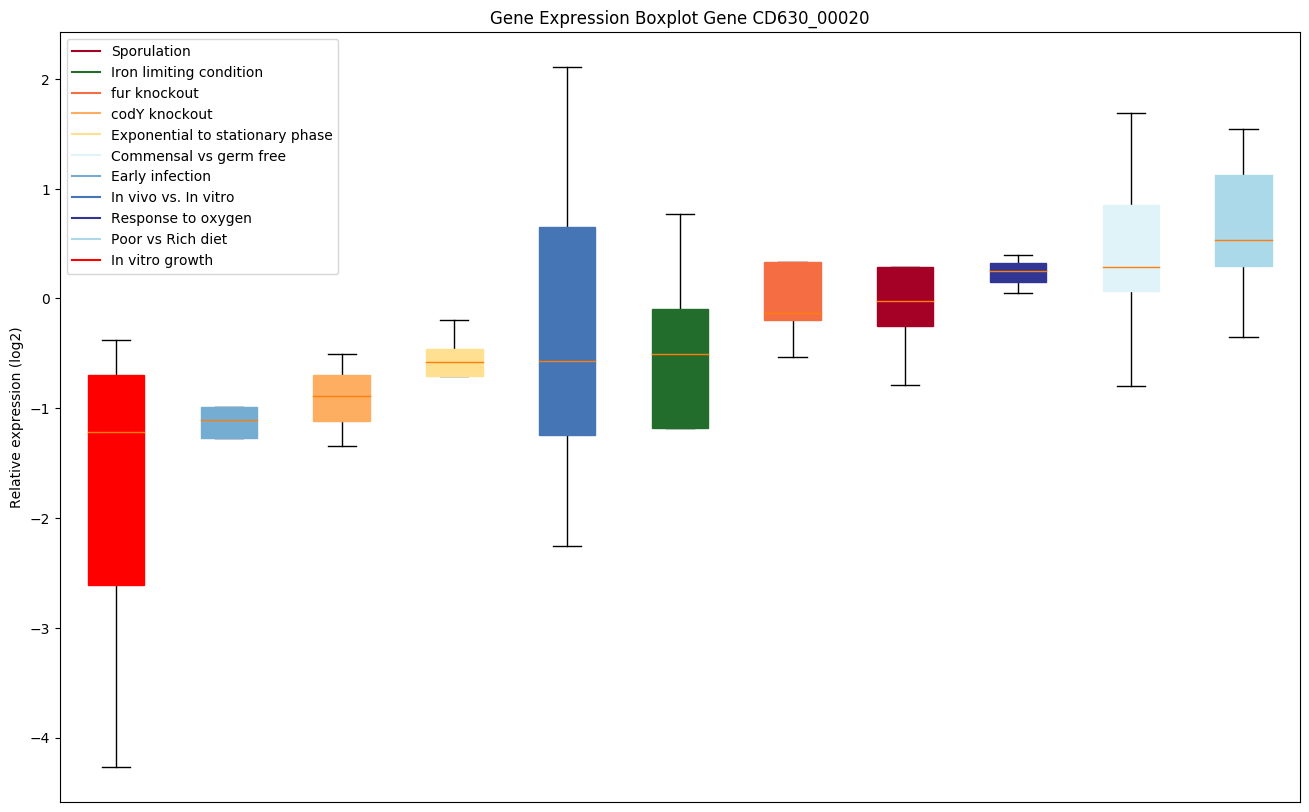 |
|||
| CD630_10760 | PTS system, mannose/fructose/sorbose IIBcomponent |
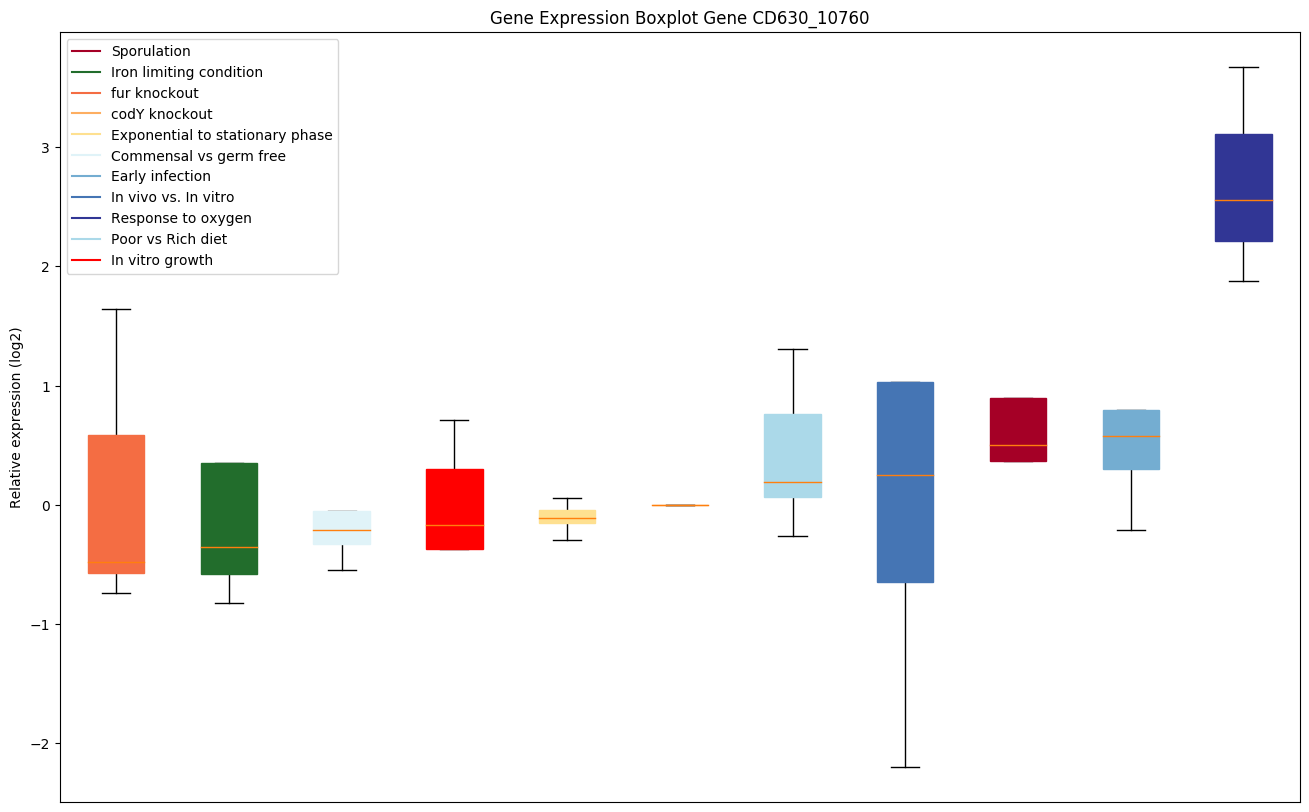 |
|||||
| CD630_17320 | Putative cobalamin-binding protein |
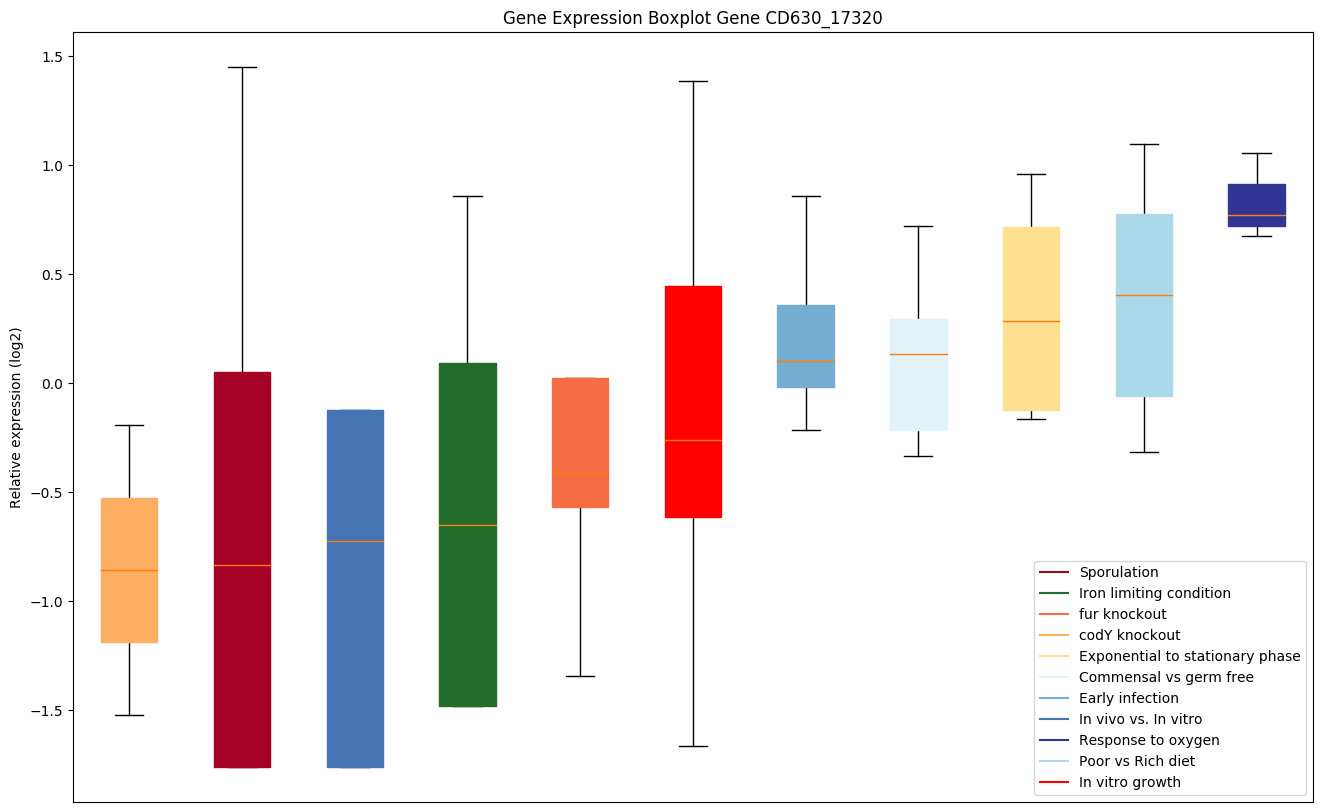 |
|||||
| CD630_00060 | gyrA | DNA gyrase subunit A | A type II topoisomerase that negatively supercoils closed circular double-stranded (ds) DNA in an ATP-dependent manner to modulate DNA topology and maintain chromosomes in an underwound state. Negative supercoiling favors strand separation, and DNA replication, transcription, recombination and repair, all of which involve strand separation. Also able to catalyze the interconversion of other topological isomers of dsDNA rings, including catenanes and knotted rings. Type II topoisomerases break and join 2 DNA strands simultaneously in an ATP-dependent manner. |
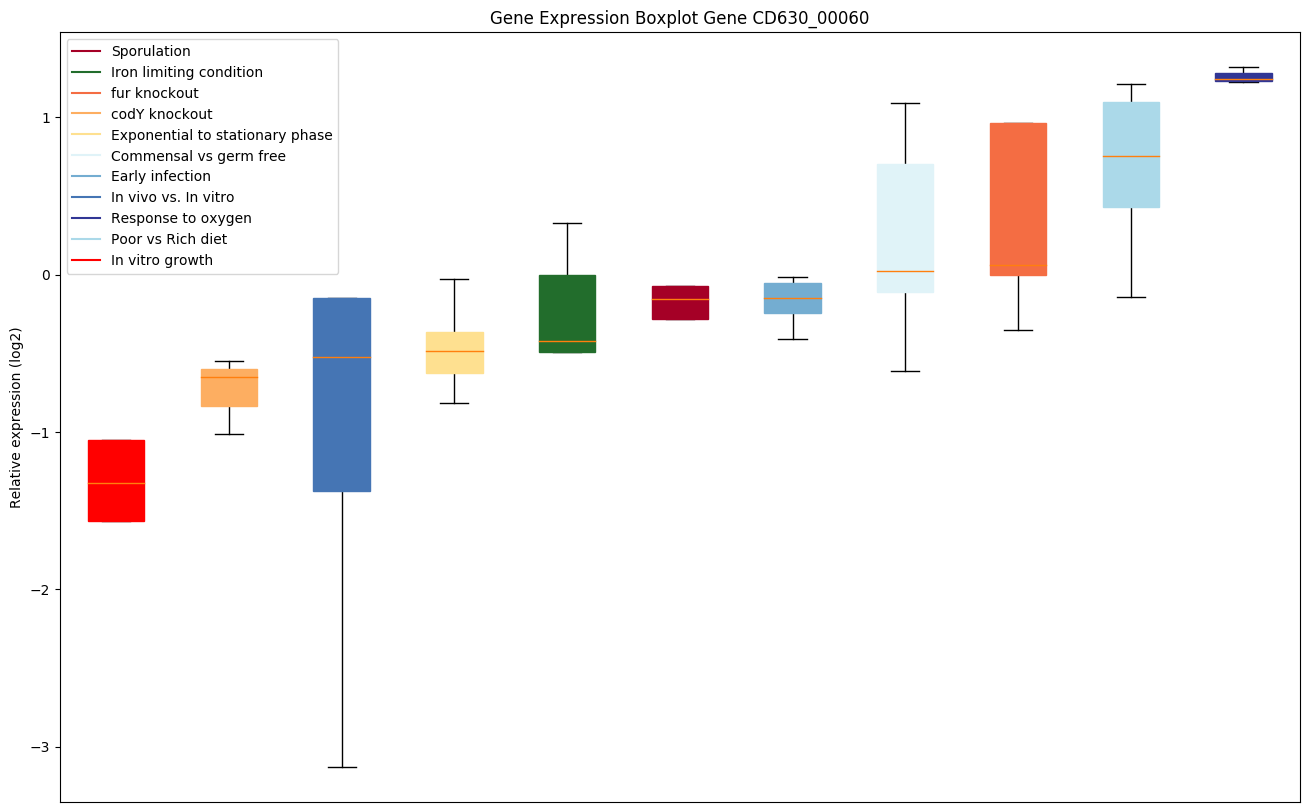 |
|||
| CD630_10740 | PTS system, mannose/fructose/sorbose IIAcomponent |
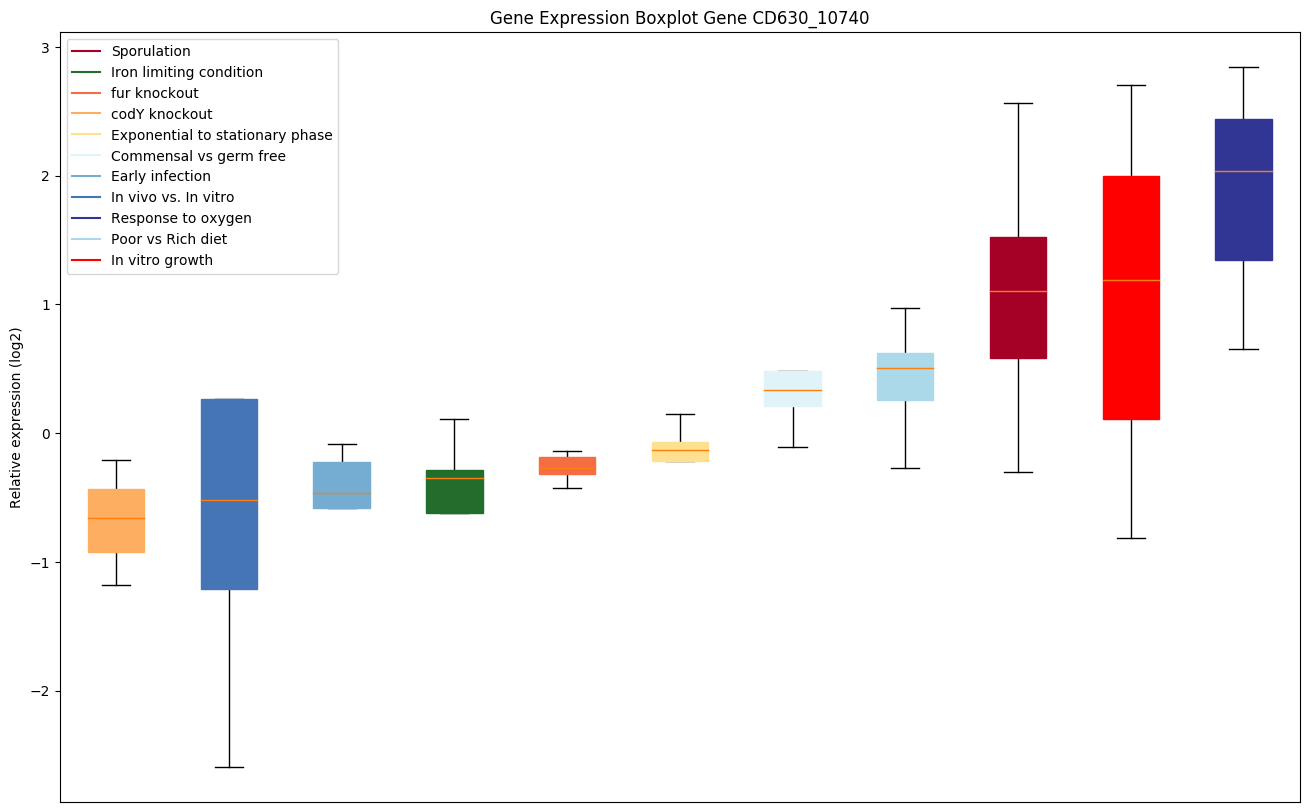 |
|||||
| CD630_17340 | Conserved hypothetical protein |
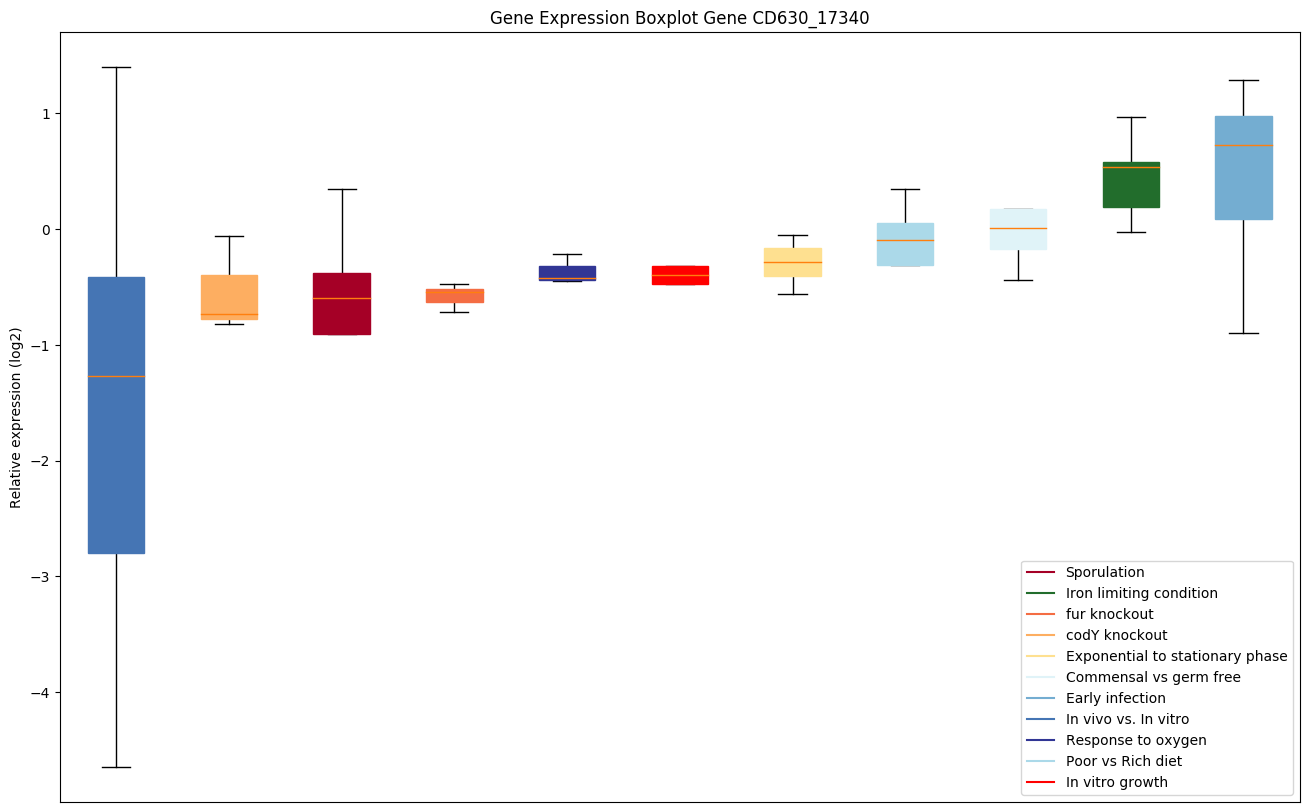 |
|||||
| CD630_06970 | Putative regulator of K+ conductance | Yes |
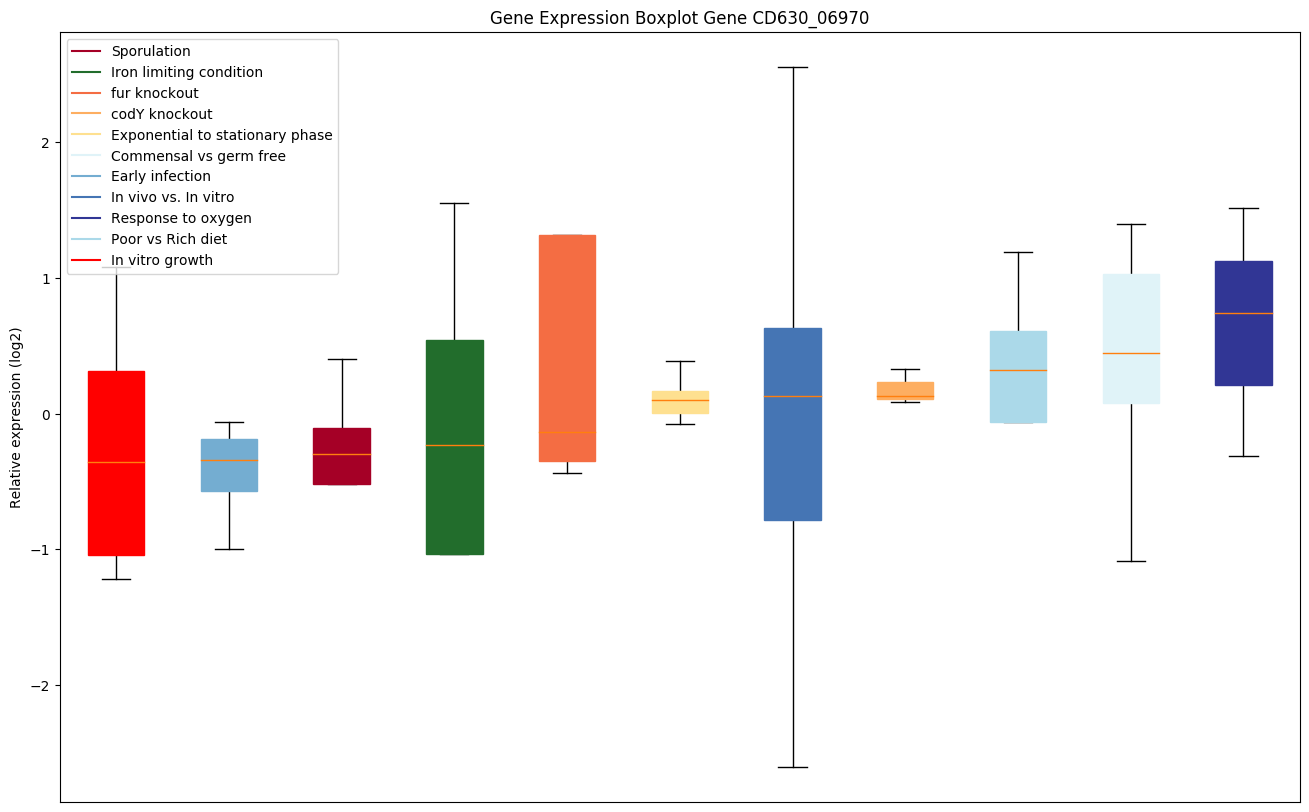 |
||||
| CD630_14080 | ddl | D-alanine--D-alanine ligase (D-alanylalaninesynthetase) (D-Ala-D-Ala ligase) | Cell wall formation. | Yes | Yes | Yes |
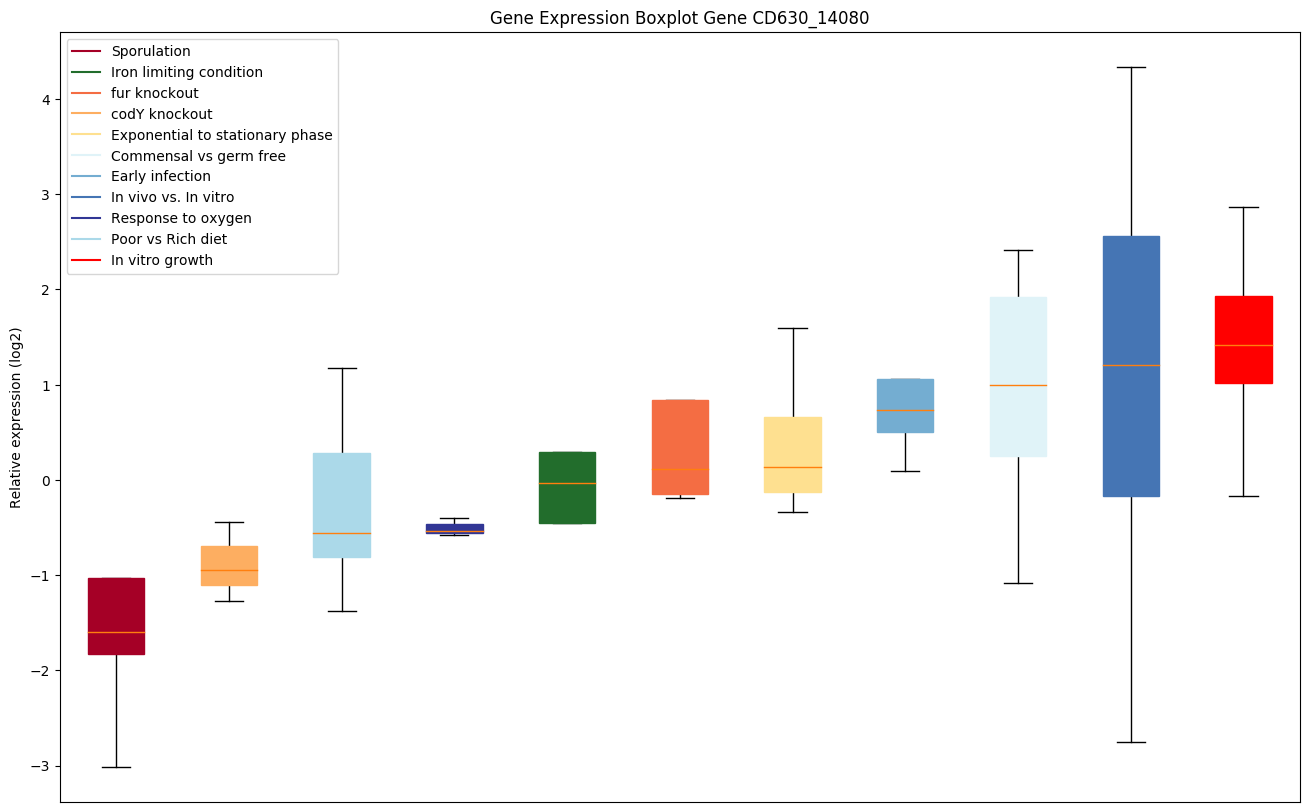 |
| CD630_14060 | Conserved hypothetical protein |
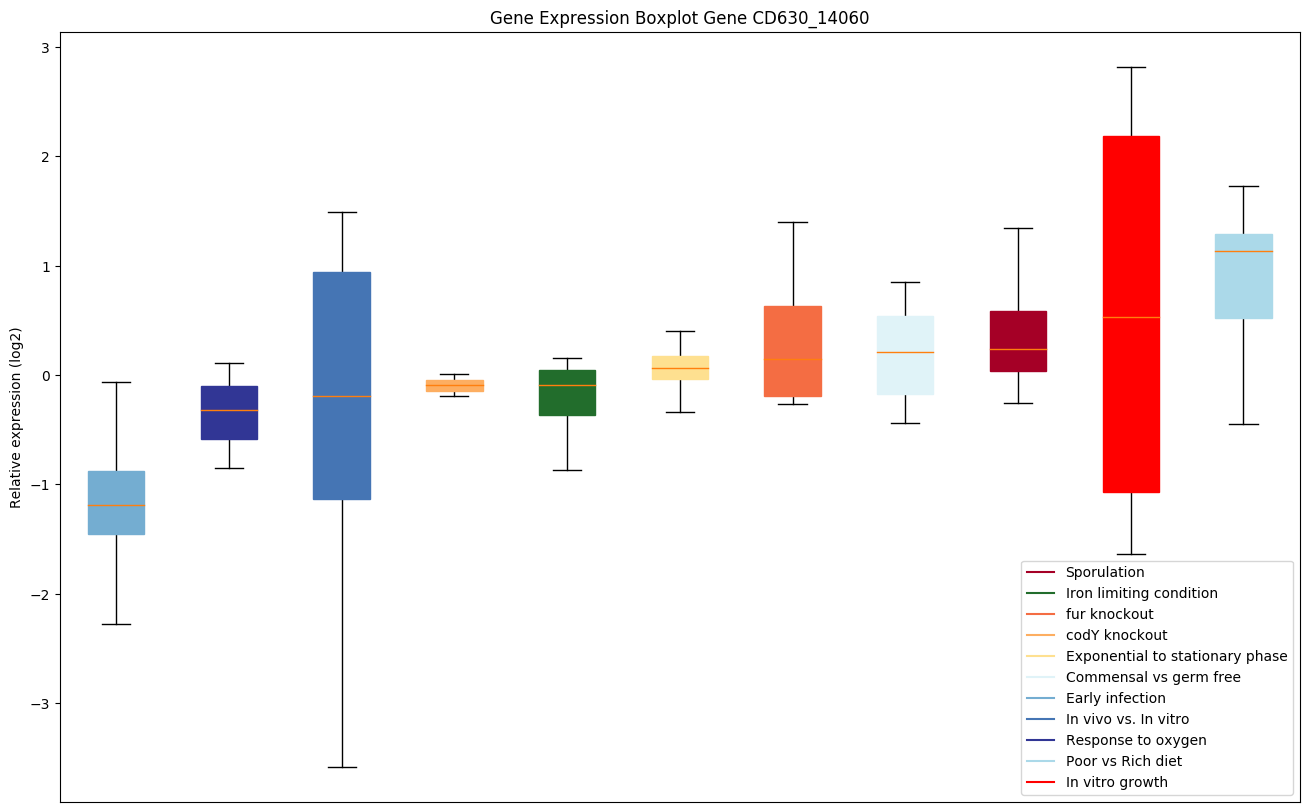 |
|||||
| CD630_23051 | Fragment of transposase (N-terminal region) |
 |
|||||
| CD630_06960 | Putative cation transport protein, Trk family | Yes |
 |
||||
| CD630_15310 | Two-component response regulator |
 |
|||||
| CD630_22630 | prsA | Peptidyl-prolyl cis-trans isomerase, PpiC-type |
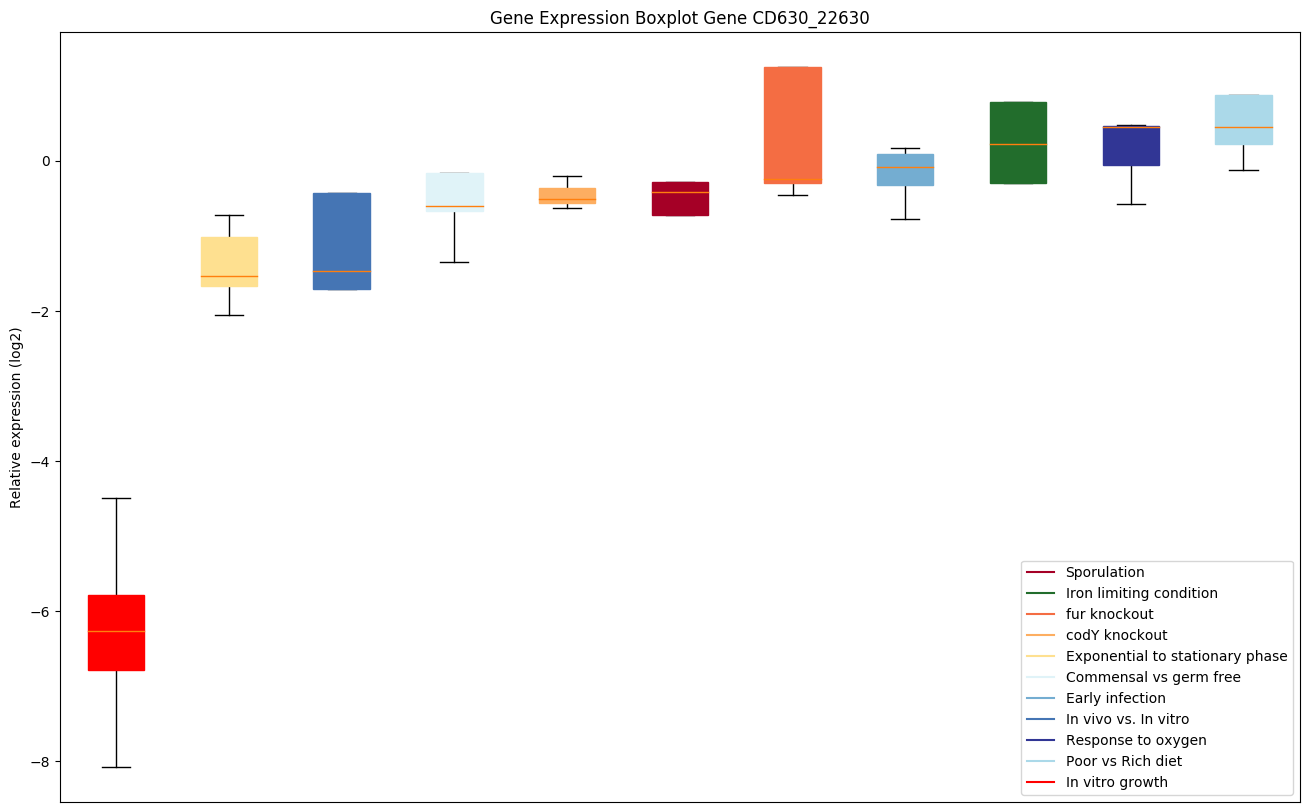 |

