Module 379
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.48 | 28 | 62 | NA | NA |
Bicluster expression profile
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
| TF | Module | Confidence Score | Beta |
|---|---|---|---|
|
Transcriptional regulator, AsnC/Lrp family |
379 | 0.19 | 0.1606 |
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
| TF | Module | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
|
prdR Transcriptional regulator, sigma-54-dependent |
379 | 7 | 0.00 | 0.00 |
The module is significantly enriched with genes associated to the indicated functional terms
The enrichment was determined with hypergeometric test using the genome functional annotation compiled by Girinathan et al (2020).
| Module | Pathway | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
| 379 | Transport Binding Proteins: Dipeptides and Oligopeptides | 6 | 0.000000 | 0.000000 |
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
| CD630_35230 | ksgA | Dimethyladenosine transferase(S-adenosylmethionine-6-N', N'-adenosyl(rRNA)dimethyltransferase) | Specifically dimethylates two adjacent adenosines (A1518 and A1519) in the loop of a conserved hairpin near the 3'-end of 16S rRNA in the 30S particle. May play a critical role in biogenesis of 30S subunits. |
 |
|||
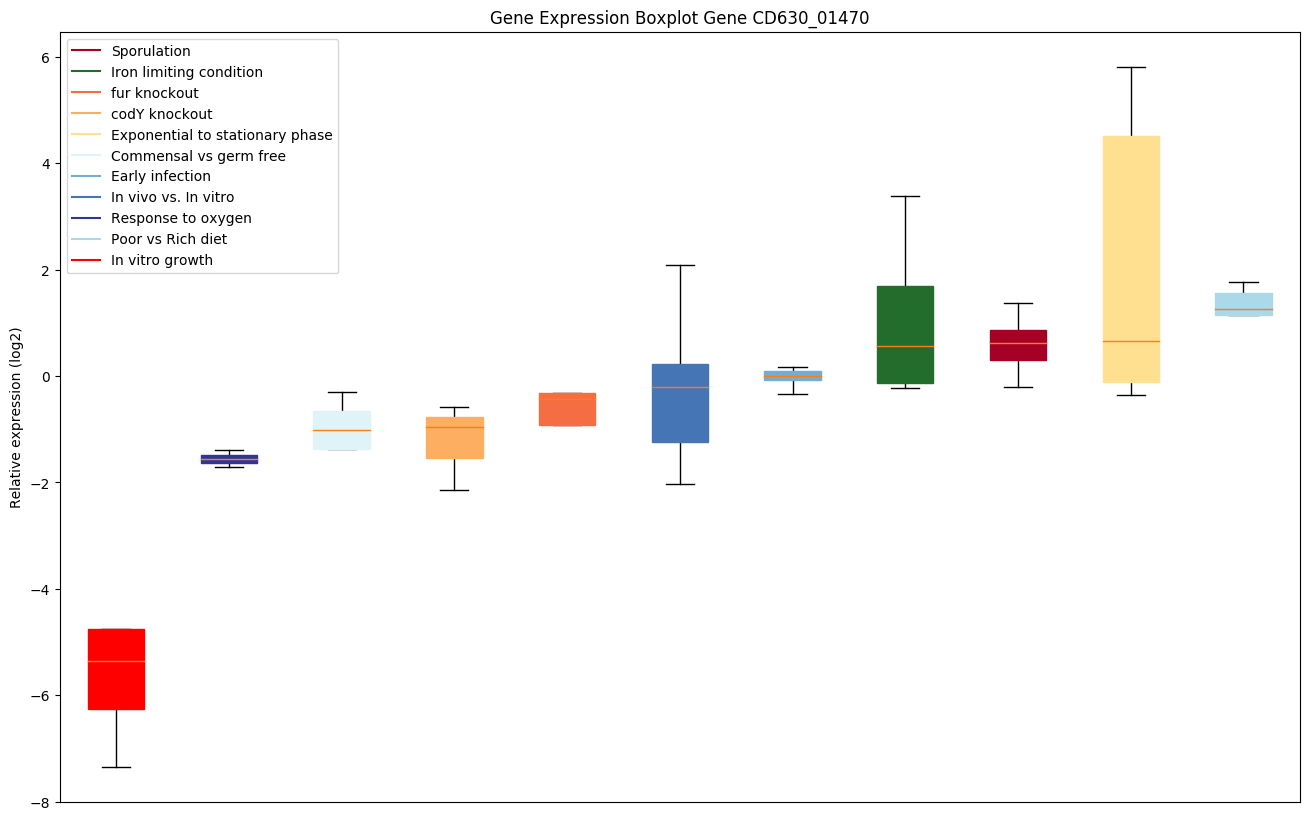
| CD630_01470 | Putative transporter | Mediates riboflavin uptake, may also transport FMN and roseoflavin. Probably a riboflavin-binding protein that interacts with the energy-coupling factor (ECF) ABC-transporter complex. Unlike classic ABC transporters this ECF transporter provides the energy necessary to transport a number of different substrates. The substrates themselves are bound by transmembrane, not extracytoplasmic soluble proteins. |
 |
||||
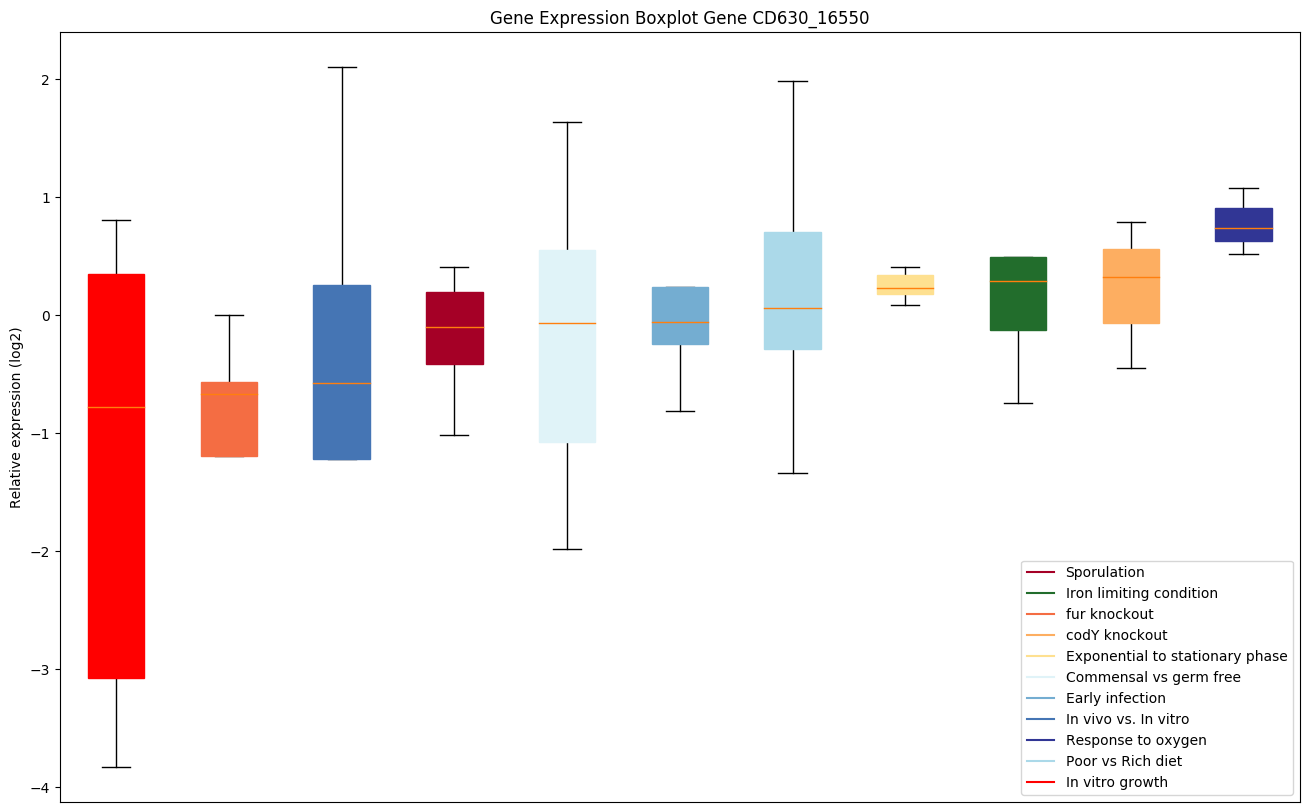
| CD630_16550 | Putative Na+/H+ antiporter NhaC-like |
 |
|||||
| CD630_09000 | opuCA | ABC-type transport system, glycinebetaine/carnitine/choline ATP-binding protein |
 |
||||
| CD630_35440 | Transcriptional regulator, AsnC/Lrp family |
 |
|||||
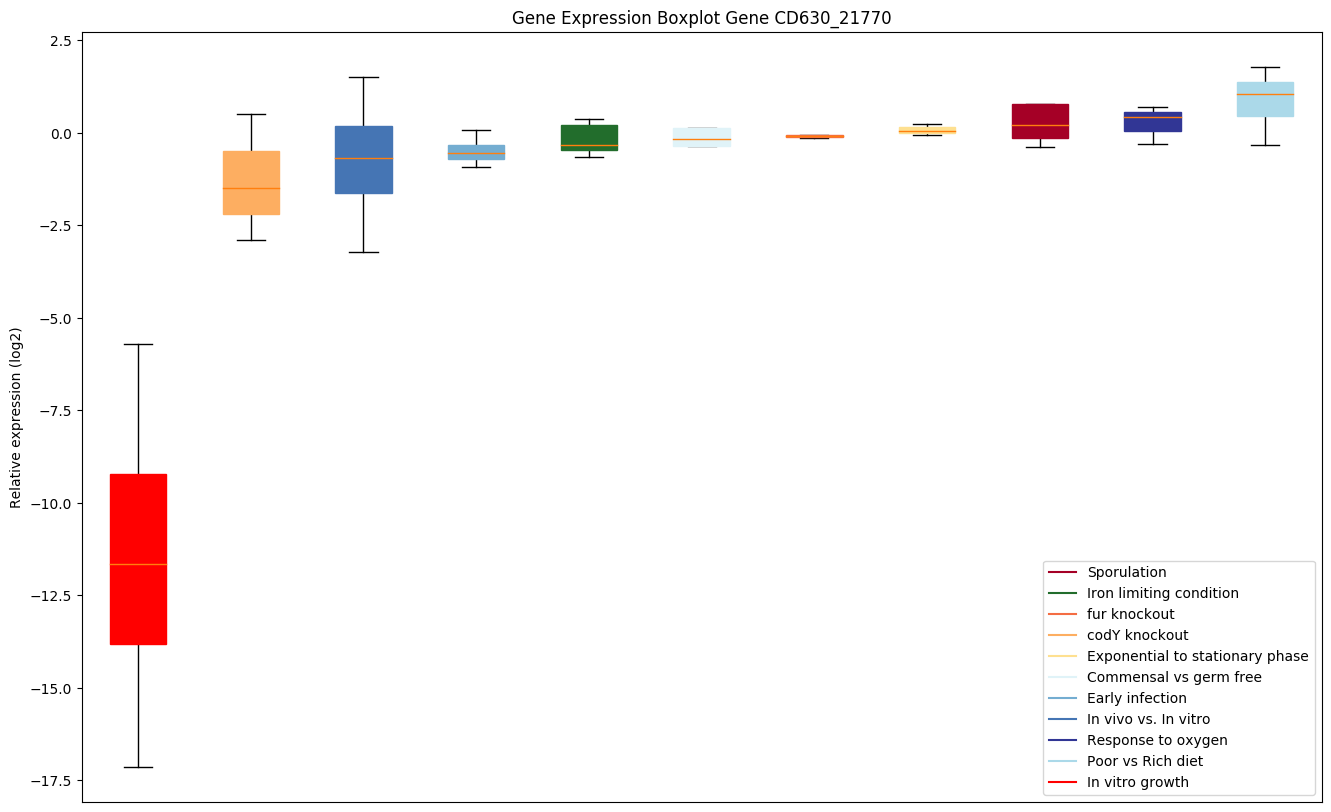
| CD630_21770 | ABC-type transport system,cystine/aminoacid-family extracellular solute-bindingprotein |
 |
|||||
| CD630_22020 | Putative Na+/solute symporter, SSS family |
 |
|||||
| CD630_15780 | Putative membrane-associated ribonuclease BN-like(Rbn) |
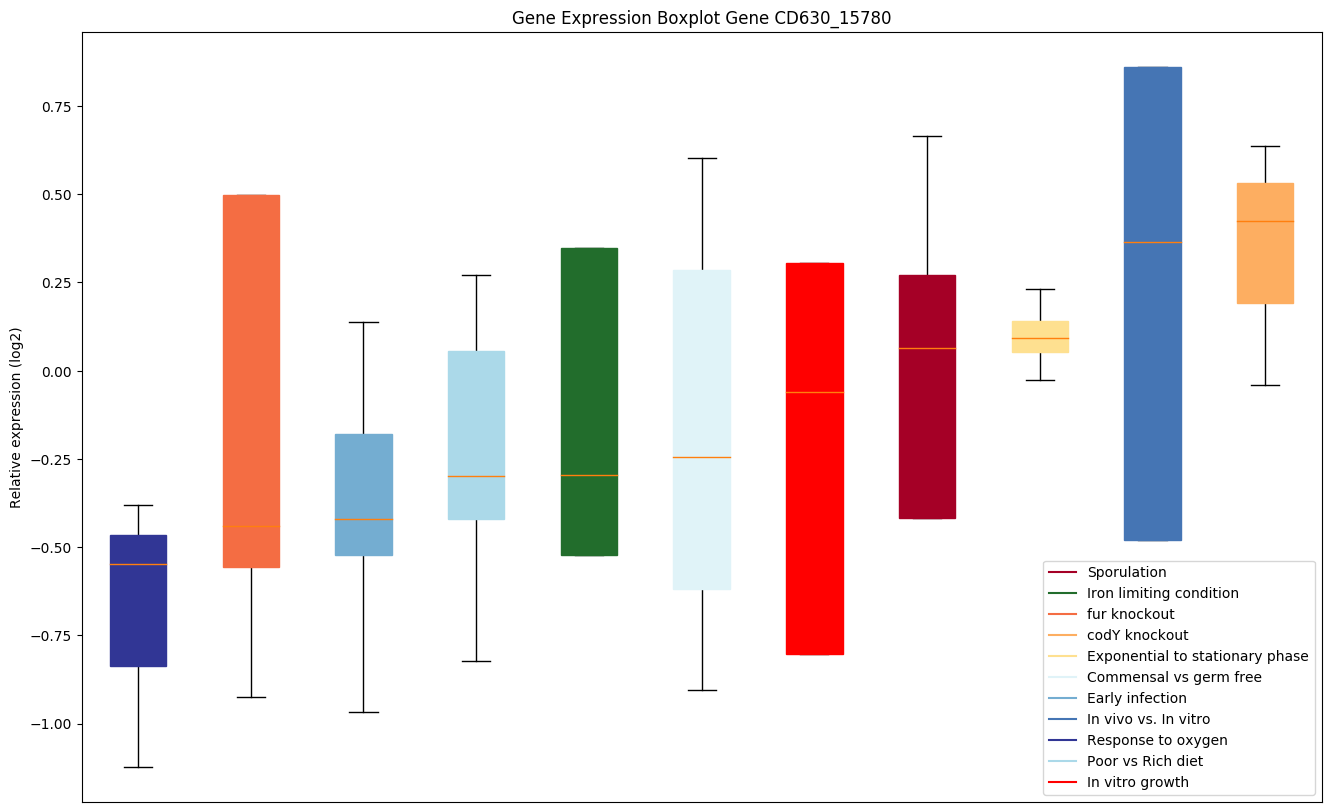 |
|||||
| CD630_05580 | glsA | Glutaminase (L-glutamine amidohydrolase) |
 |
||||
| CD630_16290 | Conserved hypothetical protein |
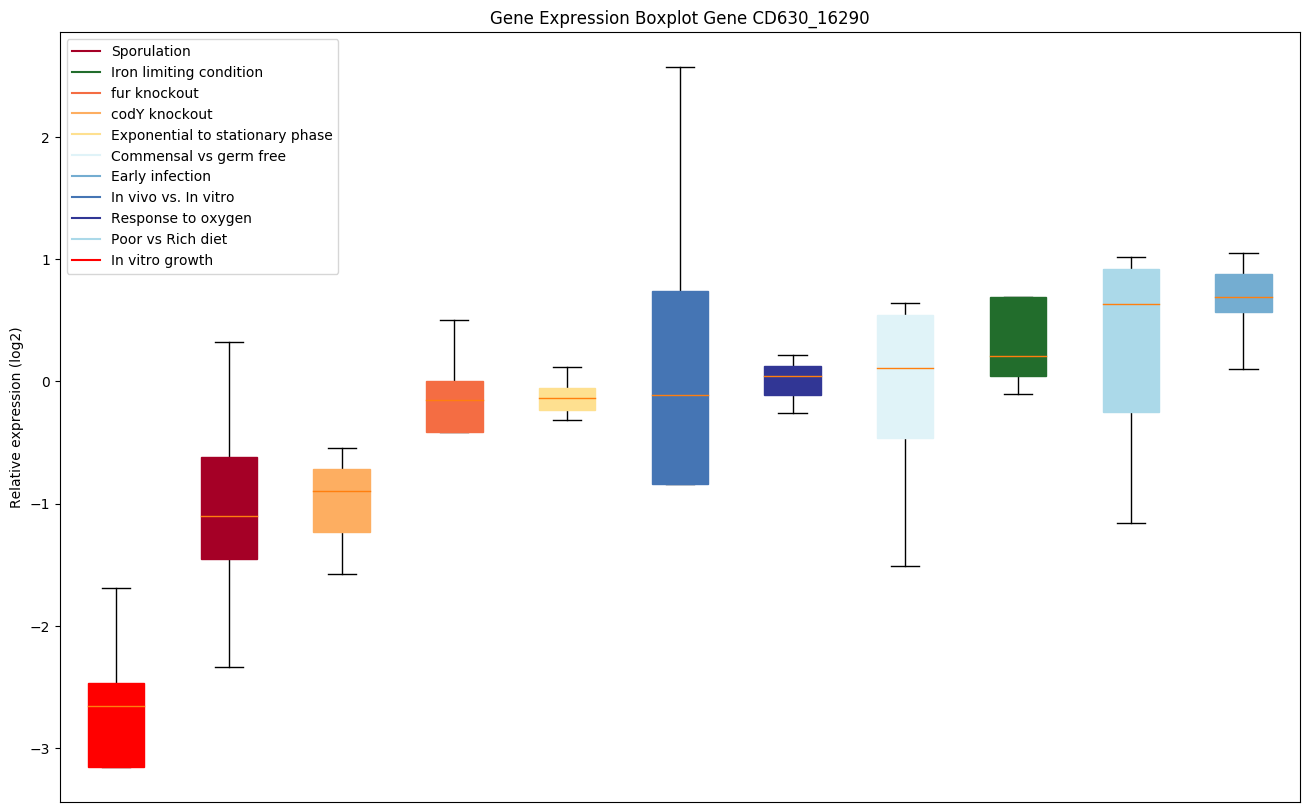 |
|||||
| CD630_25160 | ansB | L-asparaginase |
 |
||||
| CD630_21750 | ABC-type transport system,cystine/aminoacid-family permease |
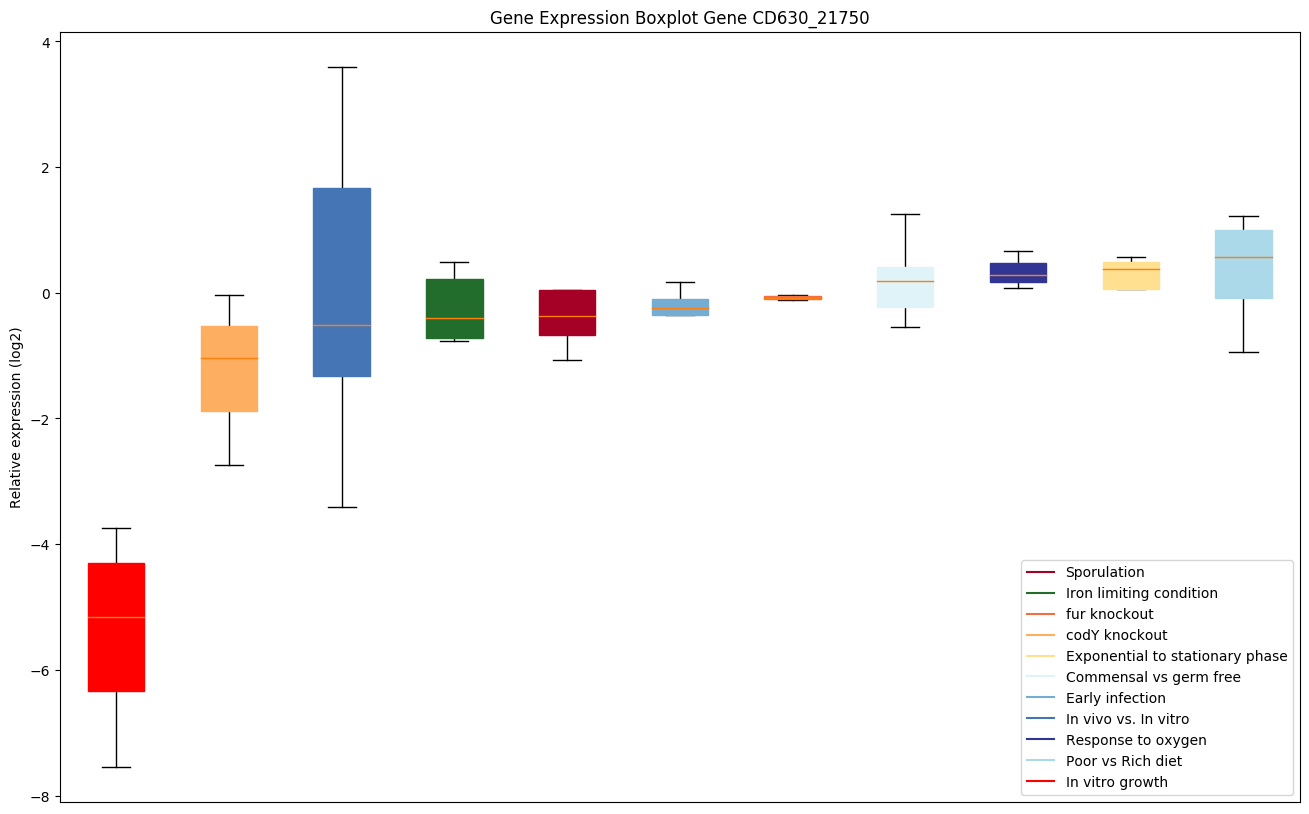 |
|||||
| CD630_21490 | Putative vancomycin resistance protein, vanWfamily |
 |
|||||
| CD630_36210 | Conserved hypothetical protein |
 |
|||||
| CD630_21730 | Putative peptidase, M20D family |
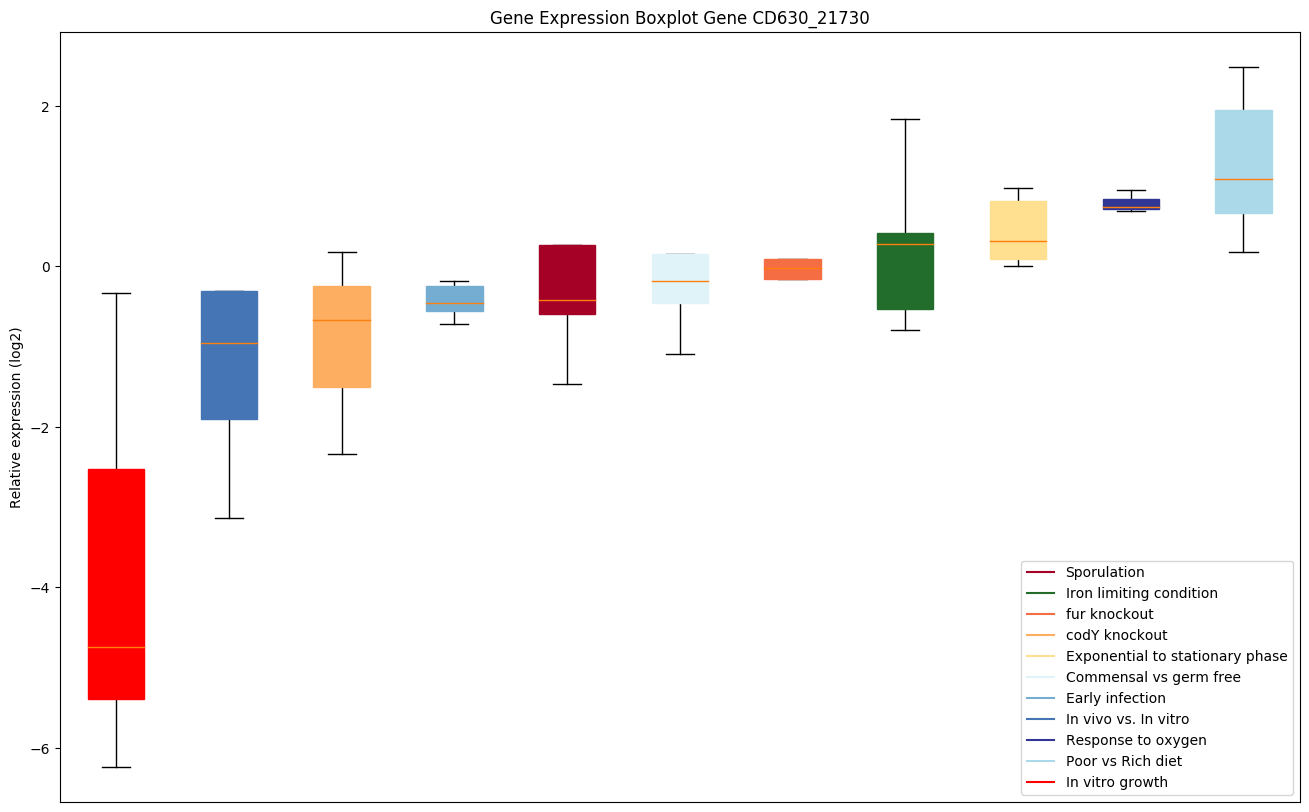 |
|||||
| CD630_21450 | DNA mismatch repair protein | Endonuclease that is involved in the suppression of homologous recombination and may therefore have a key role in the control of bacterial genetic diversity. |
 |
||||
| CD630_00210 | pycA | Pyruvate carboxylase | Catalyzes a 2-step reaction, involving the ATP-dependent carboxylation of the covalently attached biotin in the first step and the transfer of the carboxyl group to pyruvate in the second. |
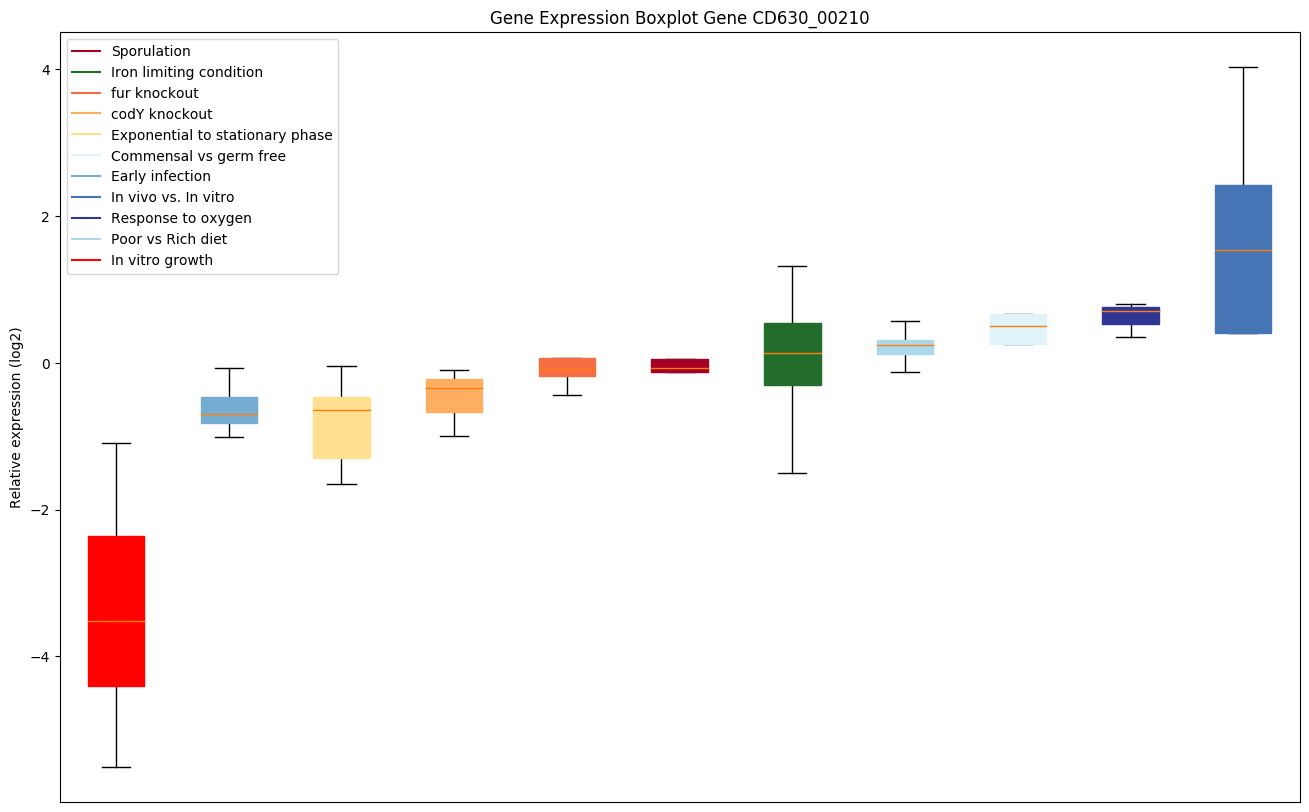 |
|||
| CD630_20480 | Transcriptional regulator, RpiR family |
 |
|||||
| CD630_35240 | Putative DNA-binding protein | Required for correct processing of both the 5' and 3' ends of 5S rRNA precursor. Cleaves both sides of a double-stranded region yielding mature 5S rRNA in one step. |
 |
||||
| CD630_09010 | opuCC | ABC-type transport system, glycinebetaine/carnitine/choline permease |
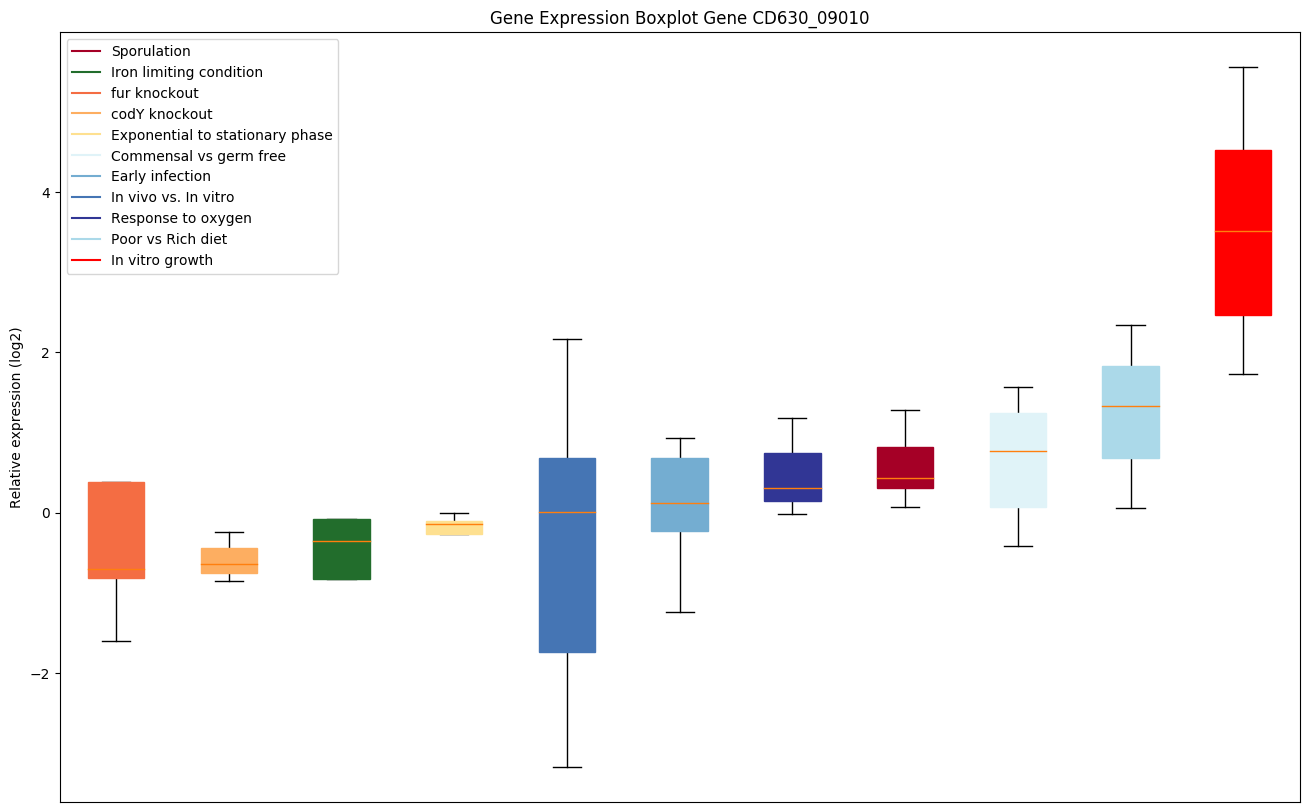 |
||||
| CD630_35620 | murI | Glutamate racemase | Provides the (R)-glutamate required for cell wall biosynthesis. | Yes |
 |
||
| CD630_35430 | Putative nicotinate phosphribosyltransferase | Catalyzes the first step in the biosynthesis of NAD from nicotinic acid, the ATP-dependent synthesis of beta-nicotinate D-ribonucleotide from nicotinate and 5-phospho-D-ribose 1-phosphate. | Yes |
 |
|||
| CD630_21760 | ABC-type transport system,cystine/aminoacid-family permease |
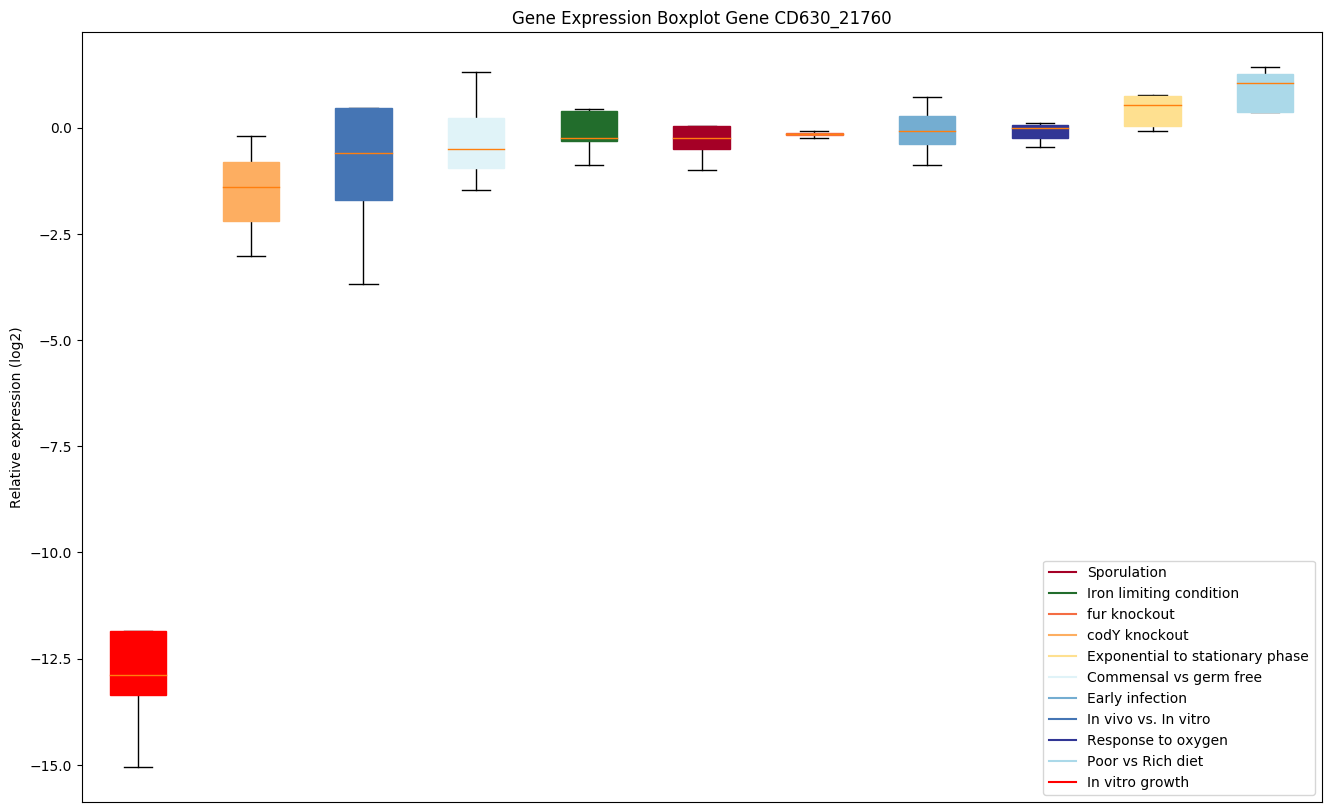 |
|||||
| CD630_22030 | Transcriptional regulator, TetR family |
 |
|||||
| CD630_21330 | Conserved hypothetical protein |
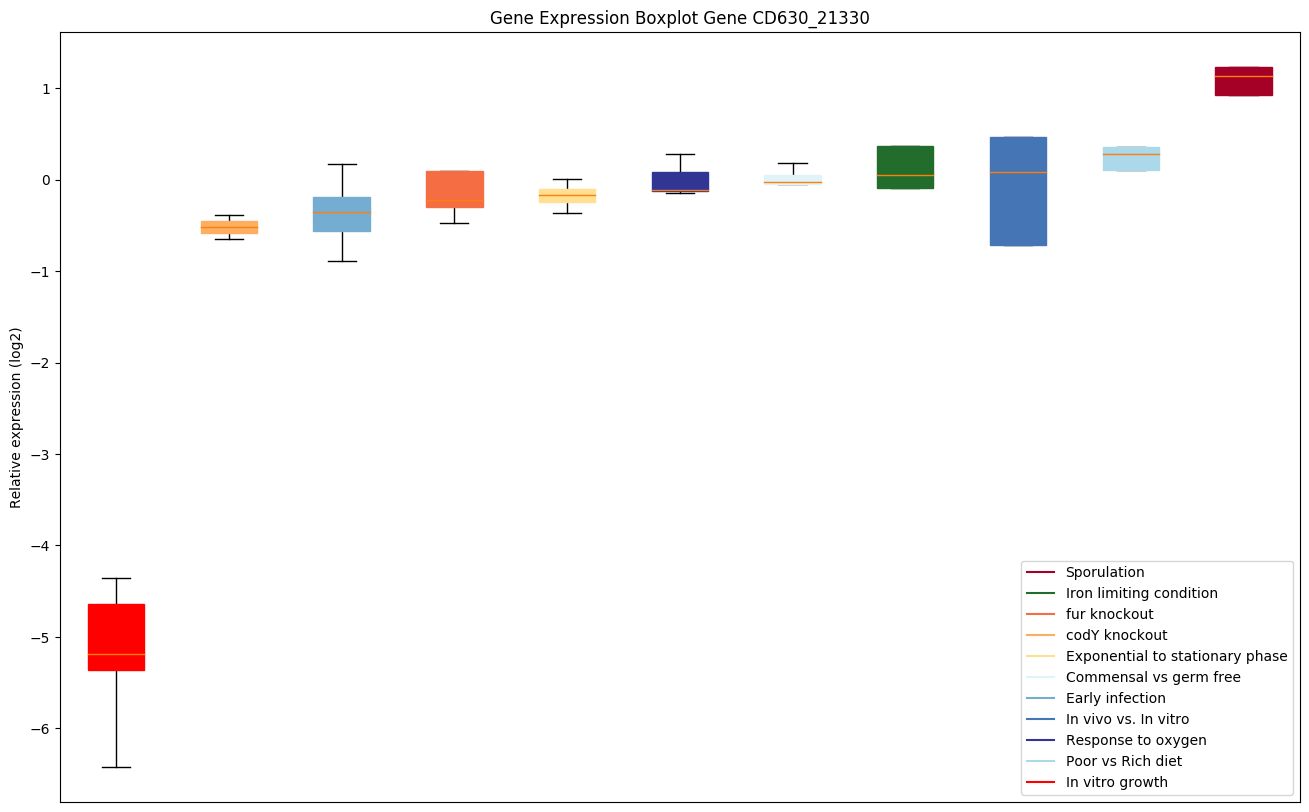 |
|||||
| CD630_21720 | ABC-type transport system,cystine/aminoacid-family ATP-binding protein |
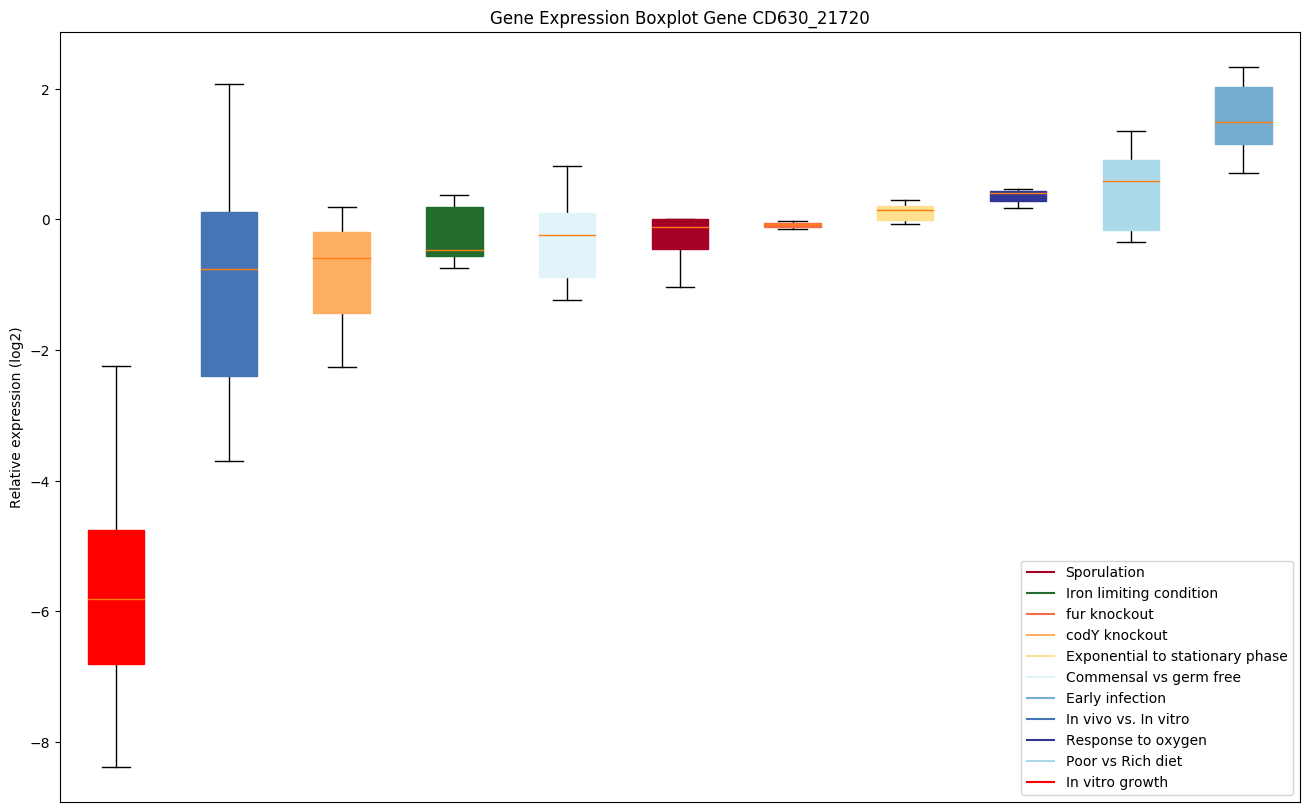 |
|||||
| CD630_05590 | Transcriptional regulator, ArsR family |
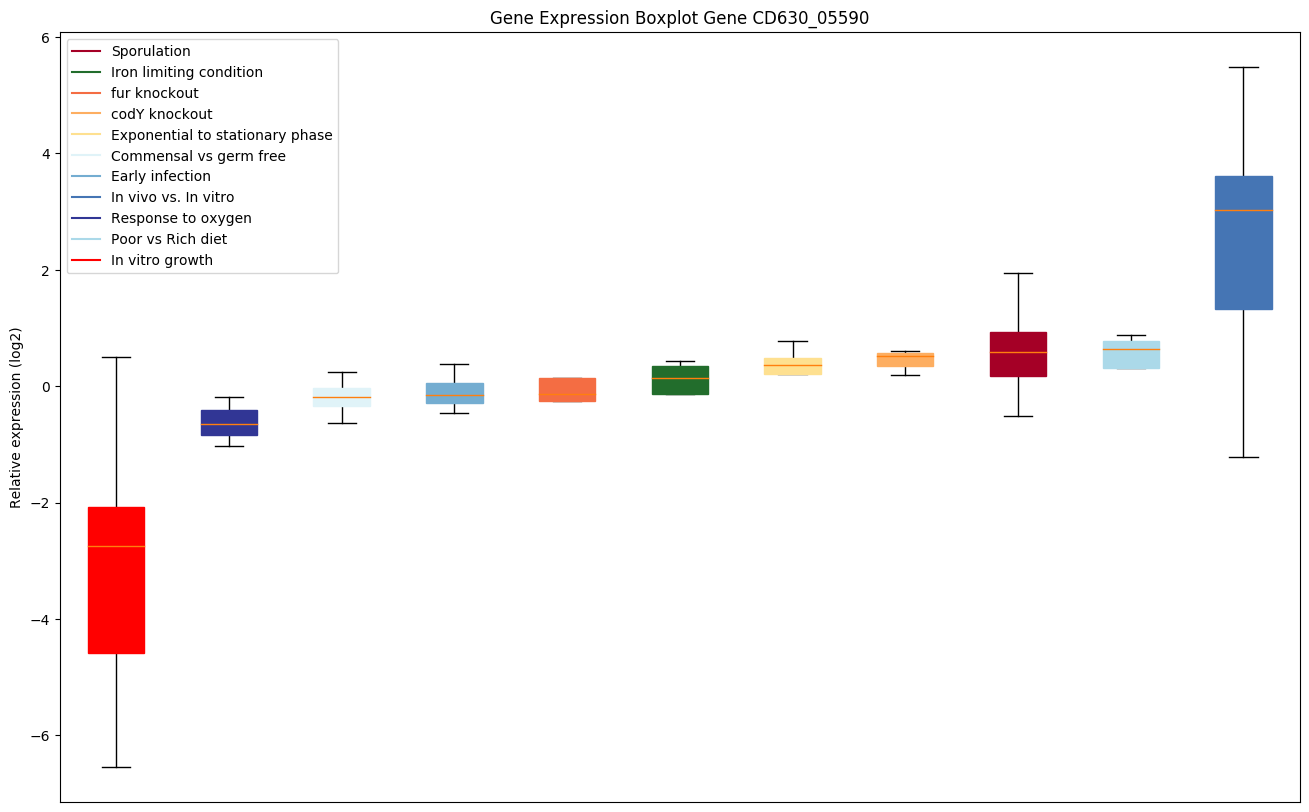 |
|||||
| CD630_21740 | ABC-type transport system,cystine/aminoacid-family extracellular solute-bindingprotein |
 |