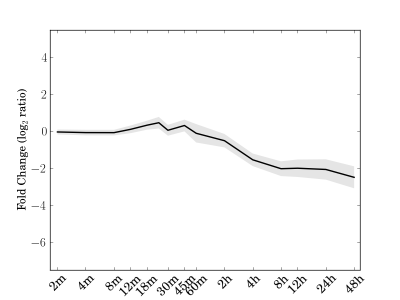
Module 172 Residual: 0.14
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0172 | v02 | 0.14 | -11.73 |
Displaying 1 - 19 of 19
| Cre01.g038400.t1.2 calreticulin 1b | Cre02.g084350.t1.2 Uncharacterized protein family (UPF0016) |
| Cre03.g144667.t1.1 ClpS-like protein | Cre03.g188700.t1.2 Plastid-lipid associated protein PAP / fibrillin family protein |
| Cre03.g207000.t1.2 abscisic acid (aba)-deficient 4 | Cre03.g209841.t1.1 |
| Cre05.g233900.t1.2 ascorbate peroxidase 4 | Cre06.g259401.t1.1 S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
| Cre06.g267600.t2.1 Lycopene beta/epsilon cyclase protein | Cre07.g342150.t1.2 Glutamyl-tRNA reductase family protein |
| Cre09.g397250.t1.2 fatty acid desaturase 5 | Cre10.g424450.t1.2 translocase of the outer mitochondrial membrane 40 |
| Cre10.g436050.t1.2 Fe superoxide dismutase 1 | Cre11.g467689.t1.1 photosynthetic electron transfer C |
| Cre12.g497300.t2.1 calcium sensing receptor | Cre12.g510050.t1.2 dicarboxylate diiron protein, putative (Crd1) |
| Cre12.g544050.t1.1 poly(A) binding protein 8 | Cre13.g588100.t1.2 rotamase cyclophilin 2 |
| Cre14.g629800.t1.2 |
|
e.value: 2.3 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: AgGAAAGaagnAAaa |
motif_0172_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 94000 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: tctGTAaTAcAtCTagGanaA |
Displaying 1 - 4 of 4
| Interaction | Weight | |
|---|---|---|
| up-regulates |
Cre09.g386753.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51 |
0.051756 |
| down-regulates |
Cre12.g523000.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51zinc finger protein 1 |
0.048563 |
| down-regulates |
Cre08.g375400.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51KNOTTED-like homeobox of Arabidopsis thaliana 7 |
0.058834 |
| up-regulates |
Cre12.g534450.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51SMAD/FHA domain-containing protein |
0.068551 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0016491 | GO:0016209 | oxidoreductase activity | antioxidant activity |

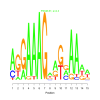
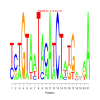
Comments