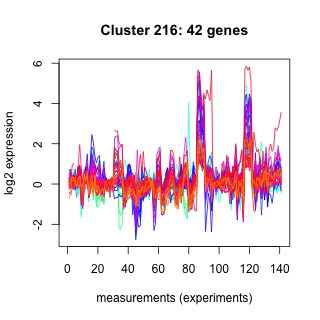
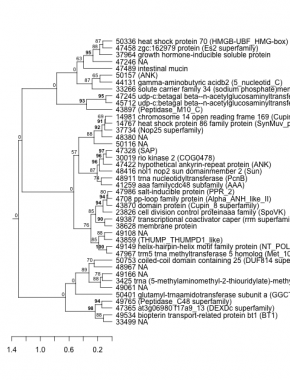
Phatr_hclust_0216 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009166 | nucleotide catabolism | 0.00886694 | 1 | Phatr_hclust_0216 |
| GO:0006817 | phosphate transport | 0.0205774 | 1 | Phatr_hclust_0216 |
| GO:0008033 | tRNA processing | 0.0577662 | 1 | Phatr_hclust_0216 |
| GO:0006260 | DNA replication | 0.0744898 | 1 | Phatr_hclust_0216 |
| GO:0006468 | protein amino acid phosphorylation | 0.0895534 | 1 | Phatr_hclust_0216 |
| GO:0006281 | DNA repair | 0.0963646 | 1 | Phatr_hclust_0216 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.143861 | 1 | Phatr_hclust_0216 |
| GO:0006396 | RNA processing | 0.1541 | 1 | Phatr_hclust_0216 |
| GO:0006412 | protein biosynthesis | 0.385887 | 1 | Phatr_hclust_0216 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.487367 | 1 | Phatr_hclust_0216 |
|
PHATRDRAFT_49149 : helix-hairpin-helix motif family protein (NT_POLXc) |
PHATRDRAFT_48416 : nol1 nop2 sun domainmember 2 (Sun) |
PHATRDRAFT_45712 : udp-c:betagal beta--n-acetylglucosaminyltransferase-like 1 (Glyco_tranf_GTA_type superfamily) |
|
|
PHATRDRAFT_49534 : biopterin transport-related protein bt1 (BT1) |
PHATRDRAFT_43859 : (THUMP_THUMPD1_like) |
PHATRDRAFT_47422 : hypothetical ankyrin-repeat protein (ANK) |
PHATRDRAFT_47245 : udp-c:betagal beta--n-acetylglucosaminyltransferase-like 1 (Glyco_tranf_GTA_type superfamily) |
|
PHATRDRAFT_47365 : at3g06980 f17a9_13 (DEXDc superfamily) |
PHATRDRAFT_30019 : rio kinase 2 (COG0478) |
PHATRDRAFT_33266 : solute carrier family 34 (sodium phosphate)member 1 (2a58) |
|
|
PHATRDRAFT_49765 : (Peptidase_C48 superfamily) |
PHATRDRAFT_38628 : membrane protein |
PHATRDRAFT_47328 : (SAP) |
PHATRDRAFT_44131 : gamma-aminobutyric acidb2 (5_nucleotid_C) |
|
PHATRDRAFT_50401 : glutamyl-trnaamidotransferase subunit a (GGCT_like superfamily) |
PHATRDRAFT_49387 : transcriptional coactivator caper (rrm superfamily) (RRM2_gar2) |
PHATRDRAFT_50157 : (ANK) |
|
|
PHATRDRAFT_23826 : cell division control proteinaaa family (SpoVK) |
PHATRDRAFT_47489 : intestinal mucin |
||
|
PHATRDRAFT_3425 : trna (5-methylaminomethyl-2-thiouridylate)-methyltransferase (tRNA_Me_trans) |
PHATRDRAFT_43870 : domain protein (Cupin_8 superfamily) |
PHATRDRAFT_37734 : (Nop25 superfamily) |
|
|
PHATRDRAFT_4708 : pp-loop family protein (Alpha_ANH_like_II) |
PHATRDRAFT_14767 : heat shock protein 86 family protein (SynMuv_product superfamily) |
PHATRDRAFT_37964 : growth hormone-inducible soluble protein |
|
|
PHATRDRAFT_47986 : salt-inducible protein (PPR_2) |
PHATRDRAFT_14981 : chromosome 14 open reading frame 169 (Cupin_8 superfamily) |
PHATRDRAFT_47458 : zgc:162979 protein (Es2 superfamily) |
|
|
PHATRDRAFT_50753 : coiled-coil domain containing 25 (DUF814 superfamily) |
PHATRDRAFT_41259 : aaa familycdc48 subfamily (AAA) |
PHATRDRAFT_43897 : (Peptidase_M10_C) |
PHATRDRAFT_50336 : heat shock protein 70 (HMGB-UBF_HMG-box) |
|
PHATRDRAFT_47967 : trm5 trna methyltransferase 5 homolog (Met_10) |
PHATRDRAFT_48911 : trna nucleotidyltransferase (PcnB) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | -0.184 | 0.286993 |
| Copper_SH | Copper_SH | 0.265 | 0.00169492 |
| Green_vs_Red_24h | Green_vs_Red_24h | 0.092 | 0.315068 |
| Silver_SH | Silver_SH | 0.155 | 0.0394977 |
| Cadmium_1.2mg | Cadmium_1.2mg | -0.042 | 0.495324 |
| BlueLight_24h | BlueLight_24h | 0.121 | 0.465909 |
| GreenLight_0.5h | GreenLight_0.5h | 0.278 | 0.325445 |
| RedLight_6h | RedLight_6h | -0.559 | 0.000694444 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 0.995848 |
| GreenLight_24h | GreenLight_24h | 0.118 | 0.391968 |
| light_6hr | light_6hr | -0.892 | 0.000561798 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | -0.671 | 0.00060241 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | -0.021 | 0.833119 |
| Ammonia_SH | Ammonia_SH | 2.401 | 0.000344828 |
| Simazine_SH | Simazine_SH | 0.528 | 0.000423729 |
| Oil_SH | Oil_SH | 0.209 | 0.0105263 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | 0.329 | 0.0021978 |
| Cadmium_SH | Cadmium_SH | 0.233 | 0.0114865 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | 0.095 | 0.406691 |
| highlight_0to6h | highlight_0to6h | 0.145 | 0.21125 |
| BlueLight_0.5h | BlueLight_0.5h | 0.257 | 0.481988 |
| Dispersant_SH | Dispersant_SH | 0.199 | 0.0218121 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | -0.131 | 0.132008 |
| light_15.5hr | light_15.5hr | -0.141 | 0.455975 |
| light_10.5hr | light_10.5hr | -0.472 | 0.00891473 |
| Dispersed_oil_SH | Dispersed_oil_SH | -0.060 | 0.465704 |
| Si_free | Si_free | 0.097 | 0.360131 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | 0.304 | 0.35598 |
| Mixture_SH | Mixture_SH | 2.211 | 0.000357143 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | 0.292 | 0.0320896 |
| light_16hr_dark_30min | light_16hr_dark_30min | -0.103 | 0.604506 |
| BlueLight_6h | BlueLight_6h | -0.267 | 0.0224044 |
| highlight_12to48h | highlight_12to48h | 0.130 | 0.412575 |
| RedLight_24h | RedLight_24h | 0.026 | 0.869376 |
| Dark_treated | Dark_treated | 0.739 | 0.0720657 |
| GreenLight_6h | GreenLight_6h | -0.246 | 0.0456818 |
| Cadmium_0.12mg | Cadmium_0.12mg | -0.038 | 0.22226 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | 0.003 | 0.969433 |
| RedLight_0.5h | RedLight_0.5h | 0.356 | 0.203448 |
| Green_vs_Red_6h | Green_vs_Red_6h | 0.313 | 0.0153061 |
| Re-illuminated_24h | Re-illuminated_24h | 0.163 | 0.103556 |