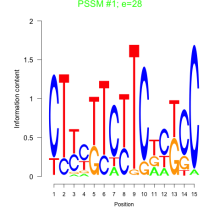
Thaps_bicluster_0012 Residual: 0.49
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0012 | 0.49 | Thalassiosira pseudonana |
Displaying 1 - 37 of 37
" class="views-fluidgrid-wrapper clear-block">
10164 hypothetical protein
10800 hypothetical protein
11026 hypothetical protein
11876 hypothetical protein
11962 hypothetical protein
1420 hypothetical protein
18351 Maf
18954 (APT2) PRTases_typeI
20223 RRM_HP0827_like
21830 WLM superfamily
22607 S_TKc
23152 G1/S-specific cyclin e2-like protein
24747 hypothetical protein
25100 PAP_fibrillin superfamily
25853 hypothetical protein
2604 hypothetical protein
261285 HSP70_NBD
261825 Tryp_SPc
262322 (Tp_Hox5b) regulator [Rayko]
263766 SBF superfamily
26467 hypothetical protein
267973 hypothetical protein
270139 Aa_trans superfamily
2880 TspO_MBR
3159 Trichoplein
33376 DPCK
36563 MYSc
38039 (Tp_Myb2R5) SANT
38815 (Rpb8) RNA_pol_Rpb8 superfamily
4350 hypothetical protein
6656 PROCN
7702 hypothetical protein
7734 hypothetical protein
7880 hypothetical protein
8166 hypothetical protein
8354 hypothetical protein
8502 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0019079 | viral genome replication | 0.00248088 | 1 | Thaps_bicluster_0012 |
| GO:0000074 | regulation of cell cycle | 0.00922038 | 1 | Thaps_bicluster_0012 |
| GO:0009116 | nucleoside metabolism | 0.0148008 | 1 | Thaps_bicluster_0012 |
| GO:0006814 | sodium ion transport | 0.0269812 | 1 | Thaps_bicluster_0012 |
| GO:0006865 | amino acid transport | 0.0696965 | 1 | Thaps_bicluster_0012 |
| GO:0006350 | transcription | 0.0720184 | 1 | Thaps_bicluster_0012 |
| GO:0006508 | proteolysis and peptidolysis | 0.157438 | 1 | Thaps_bicluster_0012 |
| GO:0016567 | protein ubiquitination | 0.179494 | 1 | Thaps_bicluster_0012 |
| GO:0006468 | protein amino acid phosphorylation | 0.41786 | 1 | Thaps_bicluster_0012 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.545155 | 1 | Thaps_bicluster_0012 |


Comments