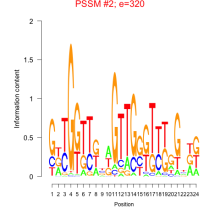
Thaps_bicluster_0046 Residual: 0.42
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0046 | 0.42 | Thalassiosira pseudonana |
Displaying 1 - 37 of 37
" class="views-fluidgrid-wrapper clear-block">
10047 hypothetical protein
10298 hypothetical protein
10603 DUF563
1118 hypothetical protein
1186 hypothetical protein
17193 HP_PGM_like
20569 NAT_SF
2088 UAA
21328 hypothetical protein
21433 hypothetical protein
21673 hypothetical protein
21732 Atx10homo_assoc superfamily
22288 hypothetical protein
22394 hypothetical protein
22601 hypothetical protein
22668 SUL1
2295 hypothetical protein
23027 hypothetical protein
23504 hypothetical protein
23770 hypothetical protein
23815 hypothetical protein
23834 hypothetical protein
24375 P4Hc
24519 hypothetical protein
24926 MATH
25656 Lactonase
261857 DHFR
263074 Synaptobrevin superfamily
3011 hypothetical protein
3037 hypothetical protein
3397 FixC
4507 hypothetical protein
5913 (Tp_bZIP10) regulator [Rayko]
7700 hypothetical protein
7725 hypothetical protein
9031 VPS28 superfamily
960 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015977 | carbon utilization by fixation of carbon dioxide | 0.0031011 | 1 | Thaps_bicluster_0046 |
| GO:0006545 | glycine biosynthesis | 0.00619321 | 1 | Thaps_bicluster_0046 |
| GO:0009165 | nucleotide biosynthesis | 0.0123506 | 1 | Thaps_bicluster_0046 |
| GO:0016192 | vesicle-mediated transport | 0.0485536 | 1 | Thaps_bicluster_0046 |
| GO:0006310 | DNA recombination | 0.0574088 | 1 | Thaps_bicluster_0046 |
| GO:0006030 | chitin metabolism | 0.0632695 | 1 | Thaps_bicluster_0046 |
| GO:0006260 | DNA replication | 0.103352 | 1 | Thaps_bicluster_0046 |
| GO:0006812 | cation transport | 0.125531 | 1 | Thaps_bicluster_0046 |
| GO:0019538 | protein metabolism | 0.133715 | 1 | Thaps_bicluster_0046 |
| GO:0006281 | DNA repair | 0.133715 | 1 | Thaps_bicluster_0046 |

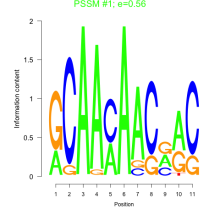
Comments