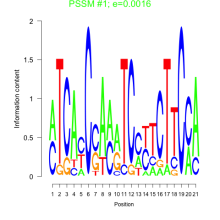
Thaps_bicluster_0080 Residual: 0.40
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0080 | 0.40 | Thalassiosira pseudonana |
Displaying 1 - 33 of 33
" class="views-fluidgrid-wrapper clear-block">
10314 COG0429
10352 hypothetical protein
11438 hypothetical protein
11490 hypothetical protein
11664 DUF3510
1467 hypothetical protein
15292 Fumble
1985 hypothetical protein
20640 (Tp_CCCH11) regulator [Rayko]
20975 hypothetical protein
2133 NBD_sugar-kinase_HSP70_actin superfamily
22437 CTNNBL superfamily
23573 hypothetical protein
24683 Raptor_N
25162 hypothetical protein
260866 (ORC2) ORC2 superfamily
262103 C2_PLC_like
263010 KU superfamily
263327 MFS_1
263562 ECM4
264833 Lig
269863 PepO
270359 hypothetical protein
30310 Aldo_ket_red
32230 (MAPK) STKc_MAPK
36582 Cullin
5393 hypothetical protein
6299 Thr-dehyd
6458 Ssu72 superfamily
6950 prc
7939 hypothetical protein
8334 hypothetical protein
9026 Phytochelatin superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015937 | coenzyme A biosynthesis | 0.00289436 | 1 | Thaps_bicluster_0080 |
| GO:0006303 | double-strand break repair via nonhomologous end-joining | 0.00289436 | 1 | Thaps_bicluster_0080 |
| GO:0007049 | cell cycle | 0.0200978 | 1 | Thaps_bicluster_0080 |
| GO:0006629 | lipid metabolism | 0.080847 | 1 | Thaps_bicluster_0080 |
| GO:0006260 | DNA replication | 0.0967972 | 1 | Thaps_bicluster_0080 |
| GO:0007165 | signal transduction | 0.109892 | 1 | Thaps_bicluster_0080 |
| GO:0007242 | intracellular signaling cascade | 0.115081 | 1 | Thaps_bicluster_0080 |
| GO:0006281 | DNA repair | 0.125374 | 1 | Thaps_bicluster_0080 |
| GO:0005975 | carbohydrate metabolism | 0.196738 | 1 | Thaps_bicluster_0080 |
| GO:0006508 | proteolysis and peptidolysis | 0.201185 | 1 | Thaps_bicluster_0080 |

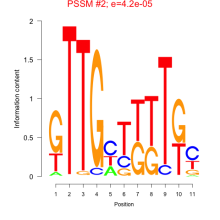
Comments