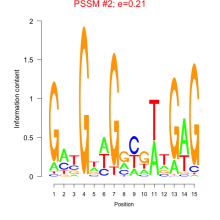
Thaps_bicluster_0100 Residual: 0.39
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0100 | 0.39 | Thalassiosira pseudonana |
Displaying 1 - 36 of 36
" class="views-fluidgrid-wrapper clear-block">
10533 hypothetical protein
11542 PseudoU_synth superfamily
12821 (Sig1) hypothetical protein
14096 (AMT3) amt
15605 PKc_like superfamily
1564 DUF667
17492 COG1179
20625 DUF4602
21346 hypothetical protein
21802 hypothetical protein
23048 hypothetical protein
23865 CE4_SF superfamily
23872 GLTP superfamily
23897 hypothetical protein
24292 (Tp_CCCH9) regulator [Rayko]
25697 ANK
262616 (MYB22)
262908 DEAD
262980 DEXDc
264799 intraflagellar transport protein, bld1, osm-6
264883 hypothetical protein
26553 ABC_MTABC3_MDL1_MDL2
268143 Nudix_Hydrolase
269503 (Tp_HSF_1e) regulator [Rayko]
31145 COG4581
31690 SpoVK
34973 DPPIV_N
38051 AAA_17 superfamily
39062 SpoU
39592 (NRT3) 2A0108
7019 mTERF superfamily
7482 hypothetical protein
7509 hypothetical protein
8503 DUF2431
9199 hypothetical protein
9818 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006003 | fructose 2,6-bisphosphate metabolism | 0.00371824 | 1 | Thaps_bicluster_0100 |
| GO:0008033 | tRNA processing | 0.0330245 | 1 | Thaps_bicluster_0100 |
| GO:0006810 | transport | 0.0434482 | 1 | Thaps_bicluster_0100 |
| GO:0006396 | RNA processing | 0.102965 | 1 | Thaps_bicluster_0100 |
| GO:0006468 | protein amino acid phosphorylation | 0.333459 | 1 | Thaps_bicluster_0100 |
| GO:0006508 | proteolysis and peptidolysis | 0.424754 | 1 | Thaps_bicluster_0100 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.446038 | 1 | Thaps_bicluster_0100 |
| GO:0008152 | metabolism | 0.546944 | 1 | Thaps_bicluster_0100 |

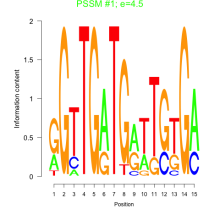
Comments