Thaps_bicluster_0163 Residual: 0.38
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0163 | 0.38 | Thalassiosira pseudonana |
Displaying 1 - 26 of 26
" class="views-fluidgrid-wrapper clear-block">
11733 hypothetical protein
16193 S49_Sppa_N_C
17695 SDR_c
1909 plant like protein
1959 hypothetical protein
19639 Got1 superfamily
21295 YCII superfamily
21327 DUF1625 superfamily
21617 Glyco_transf_15 superfamily
22246 hypothetical protein
22714 hypothetical protein
23278 GrpE
23929 hypothetical protein
25156 hypothetical protein
261882 Thioredoxin_like superfamily
262963 HMPP_kinase
264008 AdoMet_MTases superfamily
268324 60KD_IMP
270227 hypothetical protein
2779 DmpA_OAT superfamily
3126 plant_peroxidase_like superfamily
3233 Thioredoxin_like superfamily
35830 PgsA
43120 PDI_a_family
5314 hypothetical protein
8073 TauD
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009228 | thiamin biosynthesis | 0.0123506 | 1 | Thaps_bicluster_0163 |
| GO:0051205 | protein insertion into membrane | 0.0123506 | 1 | Thaps_bicluster_0163 |
| GO:0006207 | 'de novo' pyrimidine base biosynthesis | 0.0154159 | 1 | Thaps_bicluster_0163 |
| GO:0006526 | arginine biosynthesis | 0.0275887 | 1 | Thaps_bicluster_0163 |
| GO:0008654 | phospholipid biosynthesis | 0.0426071 | 1 | Thaps_bicluster_0163 |
| GO:0016192 | vesicle-mediated transport | 0.0485536 | 1 | Thaps_bicluster_0163 |
| GO:0051341 | regulation of oxidoreductase activity | 0.0690961 | 1 | Thaps_bicluster_0163 |
| GO:0006979 | response to oxidative stress | 0.0719967 | 1 | Thaps_bicluster_0163 |
| GO:0008152 | metabolism | 0.126308 | 1 | Thaps_bicluster_0163 |
| GO:0006457 | protein folding | 0.335858 | 1 | Thaps_bicluster_0163 |

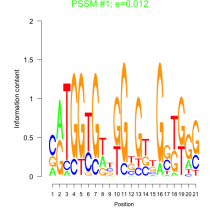

Comments