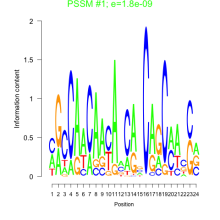
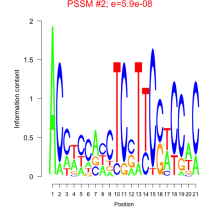
Thaps_bicluster_0025 Residual: 0.38
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0025 | 0.38 | Thalassiosira pseudonana |
Displaying 1 - 39 of 39
" class="views-fluidgrid-wrapper clear-block">
11287 COG3899
17854 HemeO
19262 hypothetical protein
21734 DnaJ-X superfamily
21813 hypothetical protein
22694 Mpv17_PMP22
22742 hypothetical protein
22874 hypothetical protein
22994 DUF1295 superfamily
23706 hypothetical protein
23902 (CLPB1) ClpB_D2-small
24157 hypothetical protein
24965 hypothetical protein
25229 Methyltransf_16 superfamily
2526 DOMON_DOH
25552 hypothetical protein
25797 (Tp_bHLH3) regulator [Rayko]
261823 FIG superfamily
26293 malate_synt_A
2674 hypothetical protein
270391 hypothetical protein
27187 (TAL1_1) PTZ00411
2895 hypothetical protein
31894 DUF4149
32029 Sterol_MT_C superfamily
3422 DUF2237 superfamily
34441 Zpr1
34507 PP2Cc
37596 SpoT
39941 PRK13357
40489 UAA
4745 hypothetical protein
5409 hypothetical protein
5993 hypothetical protein
6052 Nop superfamily
6810 MRP-L27 superfamily
8134 hypothetical protein
8673 hypothetical protein
926 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009081 | branched chain family amino acid metabolism | 0.00742955 | 1 | Thaps_bicluster_0025 |
| GO:0006097 | glyoxylate cycle | 0.00742955 | 1 | Thaps_bicluster_0025 |
| GO:0015969 | guanosine tetraphosphate metabolism | 0.0148069 | 1 | Thaps_bicluster_0025 |
| GO:0006584 | catecholamine metabolism | 0.0148069 | 1 | Thaps_bicluster_0025 |
| GO:0006098 | pentose-phosphate shunt | 0.0221325 | 1 | Thaps_bicluster_0025 |
| GO:0005975 | carbohydrate metabolism | 0.030919 | 1 | Thaps_bicluster_0025 |
| GO:0006099 | tricarboxylic acid cycle | 0.0366296 | 1 | Thaps_bicluster_0025 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0926133 | 1 | Thaps_bicluster_0025 |
| GO:0008152 | metabolism | 0.188836 | 1 | Thaps_bicluster_0025 |
| GO:0016567 | protein ubiquitination | 0.25691 | 1 | Thaps_bicluster_0025 |

Comments