Thaps_bicluster_0054 Residual: 0.44
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0054 | 0.44 | Thalassiosira pseudonana |
Displaying 1 - 36 of 36
" class="views-fluidgrid-wrapper clear-block">
11182 PLN02545
1215 E1_enzyme_family superfamily
21145 hypothetical protein
22074 hypothetical protein
22424 FKBP_C
2311 hypothetical protein
24591 hypothetical protein
24725 SEC14
25944 p450 superfamily
268931 hypothetical protein
269576 RimI
269927 DUF2453
2965 hypothetical protein
30075 (PSA5) proteasome_alpha_type_5
33310 STE14
33343 (PGD1) PLN02350
33777 S_TKc
3481 hypothetical protein
35523 ICL_PEPM
35771 PCI
36139 DUF3593
36656 DUF4336
37409 vWFA superfamily
3768 hypothetical protein
38054 hypothetical protein
38157 (FCT1) GDP-L-fucose transporter
38574 DnaJ-X superfamily
39913 (COP6) Coatomer_E
4027 hypothetical protein
40817 hypothetical protein
4186 hypothetical protein
4696 hypothetical protein
4957 hypothetical protein
5026 F1-ATPase_gamma
6718 hypothetical protein
7887 Snf7 superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006481 | C-terminal protein amino acid methylation | 0.00351458 | 1 | Thaps_bicluster_0054 |
| GO:0006097 | glyoxylate cycle | 0.00701752 | 1 | Thaps_bicluster_0054 |
| GO:0006631 | fatty acid metabolism | 0.0105089 | 1 | Thaps_bicluster_0054 |
| GO:0006098 | pentose-phosphate shunt | 0.0209137 | 1 | Thaps_bicluster_0054 |
| GO:0009401 | phosphoenolpyruvate-dependent sugar phosphotransferase system | 0.0615124 | 1 | Thaps_bicluster_0054 |
| GO:0006457 | protein folding | 0.0749473 | 1 | Thaps_bicluster_0054 |
| GO:0007018 | microtubule-based movement | 0.141059 | 1 | Thaps_bicluster_0054 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.150166 | 1 | Thaps_bicluster_0054 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.165143 | 1 | Thaps_bicluster_0054 |
| GO:0006886 | intracellular protein transport | 0.17401 | 1 | Thaps_bicluster_0054 |

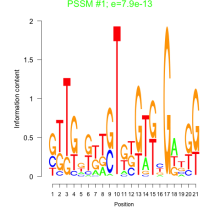

Comments