Module 19 Residual: 0.52
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0019 | v02 | 0.52 | -28.45 |
| Rv0400c Glutaryl-CoA dehydrogenase (EC 1.3.99.7) | Rv2214c Oxidoreductase, short-chain dehydrogenase/reductase family (EC 1.1.1.-) |
| Rv2215 Dihydrolipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex (EC 2.3.1.168) | Rv2216 Cell division inhibitor |
| Rv2217 Octanoate-[acyl-carrier-protein]-protein-N-octanoyltransferase | Rv2218 Lipoate synthase |
| Rv2234 Low molecular weight protein tyrosine phosphatase (EC 3.1.3.48) | Rv2235 Cytochrome oxidase biogenesis protein Surf1, facilitates heme A insertion |
| Rv3409c Cholesterol oxidase (EC 1.1.3.6) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006633 | GO:0072330 | fatty acid biosynthetic process | monocarboxylic acid biosynthetic process |
|
GO:0004361 | glutaryl-CoA dehydrogenase activity |
|
GO:0040007 | growth |
|
GO:0005829 | cytosol |
|
GO:0005886 | plasma membrane |
|
GO:0005887 | integral to plasma membrane |
|
GO:0004149 | dihydrolipoyllysine-residue succinyltransferase activity |
|
GO:0005515 | protein binding |
|
GO:0009405 | pathogenesis |
|
GO:0031405 | lipoic acid binding |
|
GO:0045254 | pyruvate dehydrogenase complex |
|
GO:0045454 | cell redox homeostasis |
|
GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity |
|
GO:0003824 | catalytic activity |
|
GO:0006464 | cellular protein modification process |
|
GO:0009107 | lipoate biosynthetic process |
|
GO:0033819 | lipoyl(octanoyl) transferase activity |
|
GO:0005506 | iron ion binding |
|
GO:0004725 | protein tyrosine phosphatase activity |
|
GO:0005576 | extracellular region |
|
GO:0006470 | protein dephosphorylation |
|
GO:0052046 | modification by symbiont of host morphology or physiology via secreted substance |
|
GO:0052083 | negative regulation by symbiont of host cell-mediated immune response |
|
GO:0004438 | phosphatidylinositol-3-phosphatase activity |
|
GO:0005618 | cell wall |
|
GO:0044119 | growth of symbiont in host cell |











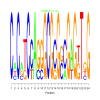
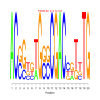
Discussion