Module 160 Residual: 0.59
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0160 | v02 | 0.59 | -7.14 |
| Rv0224c POSSIBLE METHYLTRANSFERASE (METHYLASE) | Rv0494 Lactate-responsive regulator LldR in Enterobacteria, GntR family |
| Rv0998 cyclic nucleotide-binding protein | Rv1374c |
| Rv1776c Transcriptional regulator, TetR family | Rv2771c Multimeric flavodoxin WrbA |
| Rv2816c CRISPR-associated protein Cas2 | Rv2818c CRISPR-associated protein Csm6 |
| Rv2824c CRISPR repeat RNA endoribonuclease Cas6 | Rv3097c (MTCY164.08c), len: 436. Esterase/lipase, identical to TR:P77909 (EMBL:U76006) ESTERASE/LIPASE (EC 3.1.1.3) (TRIACYLGLYCEROL LIPASE) (437 aa); N-terminal similar to N-terminal of M. tuberculosis PE family members, eg Y03A_MYCTU Q10637 hypothetical glycine-rich 49.6 kd pro (603 aa), fasta scores, opt: 334 E(): 4.8e-13, (43.4% identity in 152 aa overlap). Other relatives include MTCY1A11.25c, 7.8e-14; MTCY21B4.13c, 2.4e-12; MTCY270.06, 5.4e-12; MTCY359.33, 1.2e-11; MTC1A11.04, 1.2ee-11; cY130.seq, 1.3e-11 |
| Rv0492c GMC-type oxidoreductase | Rv0493c |
| Rv1402 Helicase PriA essential for oriC/DnaA-independent DNA replication | Rv2748c Cell division protein FtsK |
| Rv2776c Vanillate O-demethylase oxidoreductase (EC 1.14.13.-) | Rv2914c Transmembrane serine/threonine-protein kinase I PknI (EC 2.7.11.1) |
|
GO:0040007 | growth |
|
GO:0005886 | plasma membrane |
|
GO:0004806 | triglyceride lipase activity |
|
GO:0005618 | cell wall |
|
GO:0016042 | lipid catabolic process |
|
GO:0042594 | response to starvation |
|
GO:0005829 | cytosol |
|
GO:0008150 | biological_process |
|
GO:0005576 | extracellular region |
|
GO:0004672 | protein kinase activity |
|
GO:0018105 | peptidyl-serine phosphorylation |
|
GO:0018107 | peptidyl-threonine phosphorylation |
|
GO:0030145 | manganese ion binding |
|
GO:0046777 | protein autophosphorylation |


















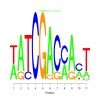

Discussion