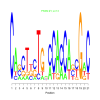
Module 198 Residual: 0.47
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0198 | v02 | 0.47 | -13.26 |
| Rv0036c | Rv0066c Isocitrate dehydrogenase [NADP] (EC 1.1.1.42); Monomeric isocitrate dehydrogenase [NADP] (EC 1.1.1.42) |
| Rv0548c Naphthoate synthase (EC 4.1.3.36) | Rv1932 Thiol peroxidase, Tpx-type (EC 1.11.1.15) |
| Rv2101 Helicase, SNF2/RAD54 family | Rv2102 |
| Rv3775 Beta-lactamase | Rv1911c Phosphatidylethanolamine-binding protein |
| Rv3774 Enoyl-CoA hydratase |
|
GO:0005886 | plasma membrane |
|
GO:0000287 | magnesium ion binding |
|
GO:0004448 | isocitrate dehydrogenase activity |
|
GO:0005576 | extracellular region |
|
GO:0005829 | cytosol |
|
GO:0042803 | protein homodimerization activity |
|
GO:0008935 | 1,4-dihydroxy-2-naphthoyl-CoA synthase activity |
|
GO:0034214 | protein hexamerization |
|
GO:0051260 | protein homooligomerization |
|
GO:0004601 | peroxidase activity |
|
GO:0051920 | peroxiredoxin activity |
|
GO:0055114 | oxidation-reduction process |
|
GO:0070402 | NADPH binding |
|
GO:0005618 | cell wall |











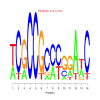
Discussion